Rejuvenating aged microglia by p16ink4a-siRNA-loaded nanoparticles increases amyloid-β clearance in animal models of Alzheimer's disease
- PMID: 38493185
- PMCID: PMC10943801
- DOI: 10.1186/s13024-024-00715-x
Rejuvenating aged microglia by p16ink4a-siRNA-loaded nanoparticles increases amyloid-β clearance in animal models of Alzheimer's disease
Abstract
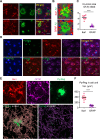
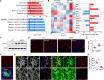
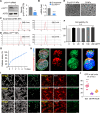
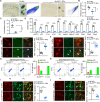
Age-dependent accumulation of amyloid plaques in patients with sporadic Alzheimer's disease (AD) is associated with reduced amyloid clearance. Older microglia have a reduced ability to phagocytose amyloid, so phagocytosis of amyloid plaques by microglia could be regulated to prevent amyloid accumulation. Furthermore, considering the aging-related disruption of cell cycle machinery in old microglia, we hypothesize that regulating their cell cycle could rejuvenate them and enhance their ability to promote more efficient amyloid clearance. First, we used gene ontology analysis of microglia from young and old mice to identify differential expression of cyclin-dependent kinase inhibitor 2A (p16ink4a), a cell cycle factor related to aging. We found that p16ink4a expression was increased in microglia near amyloid plaques in brain tissue from patients with AD and 5XFAD mice, a model of AD. In BV2 microglia, small interfering RNA (siRNA)-mediated p16ink4a downregulation transformed microglia with enhanced amyloid phagocytic capacity through regulated the cell cycle and increased cell proliferation. To regulate microglial phagocytosis by gene transduction, we used poly (D,L-lactic-co-glycolic acid) (PLGA) nanoparticles, which predominantly target microglia, to deliver the siRNA and to control microglial reactivity. Nanoparticle-based delivery of p16ink4a siRNA reduced amyloid plaque formation and the number of aged microglia surrounding the plaque and reversed learning deterioration and spatial memory deficits. We propose that downregulation of p16ink4a in microglia is a promising strategy for the treatment of Alzheimer's disease.
Keywords: Alzheimer’s disease; Cell cycle; Microglia senescence; Phagocytosis; p16ink4a.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures

References
-
- Madhavan A, Schwarz CG, Duffy JR, Strand EA, Machulda MM, Drubach DA, Kantarci K, Przybelski SA, Reid RI, Senjem ML, et al. Characterizing White Matter Tract Degeneration in Syndromic Variants of Alzheimer's Disease: A Diffusion Tensor Imaging Study. J Alzheimers Dis. 2016;49:633–643. doi: 10.3233/JAD-150502. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

