Sequence-structure-function characterization of the emerging tetracycline destructase family of antibiotic resistance enzymes
- PMID: 38493211
- PMCID: PMC10944477
- DOI: 10.1038/s42003-024-06023-w
Sequence-structure-function characterization of the emerging tetracycline destructase family of antibiotic resistance enzymes
Abstract
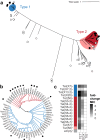
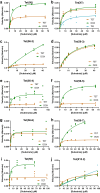
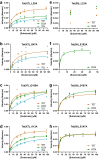
Tetracycline destructases (TDases) are flavin monooxygenases which can confer resistance to all generations of tetracycline antibiotics. The recent increase in the number and diversity of reported TDase sequences enables a deep investigation of the TDase sequence-structure-function landscape. Here, we evaluate the sequence determinants of TDase function through two complementary approaches: (1) constructing profile hidden Markov models to predict new TDases, and (2) using multiple sequence alignments to identify conserved positions important to protein function. Using the HMM-based approach we screened 50 high-scoring candidate sequences in Escherichia coli, leading to the discovery of 13 new TDases. The X-ray crystal structures of two new enzymes from Legionella species were determined, and the ability of anhydrotetracycline to inhibit their tetracycline-inactivating activity was confirmed. Using the MSA-based approach we identified 31 amino acid positions 100% conserved across all known TDase sequences. The roles of these positions were analyzed by alanine-scanning mutagenesis in two TDases, to study the impact on cell and in vitro activity, structure, and stability. These results expand the diversity of TDase sequences and provide valuable insights into the roles of important residues in TDases, and flavin monooxygenases more broadly.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
C10-Benzoate Esters of Anhydrotetracycline Inhibit Tetracycline Destructases and Recover Tetracycline Antibacterial Activity.ACS Infect Dis. 2025 Mar 14;11(3):738-749. doi: 10.1021/acsinfecdis.4c00912. Epub 2025 Feb 6. ACS Infect Dis. 2025. PMID: 39912785 Free PMC article.
-
Structure of anhydrotetracycline-bound Tet(X6) reveals the mechanism for inhibition of type 1 tetracycline destructases.Commun Biol. 2023 Apr 17;6(1):423. doi: 10.1038/s42003-023-04792-4. Commun Biol. 2023. PMID: 37062778 Free PMC article.
-
Structure-Based Design of Bisubstrate Tetracycline Destructase Inhibitors That Block Flavin Redox Cycling.J Med Chem. 2023 Mar 23;66(6):3917-3933. doi: 10.1021/acs.jmedchem.2c01629. Epub 2023 Mar 6. J Med Chem. 2023. PMID: 36877173 Free PMC article.
-
Emerging High-Level Tigecycline Resistance: Novel Tetracycline Destructases Spread via the Mobile Tet(X).Bioessays. 2020 Aug;42(8):e2000014. doi: 10.1002/bies.202000014. Epub 2020 Jun 22. Bioessays. 2020. PMID: 32567703 Review.
-
Tetracycline-Inactivating Enzymes.Front Microbiol. 2018 May 30;9:1058. doi: 10.3389/fmicb.2018.01058. eCollection 2018. Front Microbiol. 2018. PMID: 29899733 Free PMC article. Review.
Cited by
-
C10-Benzoate Esters of Anhydrotetracycline Inhibit Tetracycline Destructases and Recover Tetracycline Antibacterial Activity.ACS Infect Dis. 2025 Mar 14;11(3):738-749. doi: 10.1021/acsinfecdis.4c00912. Epub 2025 Feb 6. ACS Infect Dis. 2025. PMID: 39912785 Free PMC article.
-
Binding assays enable discovery of Tet(X) inhibitors that combat tetracycline destructase resistance.Chem Sci. 2025 May 7;16(22):9691-9704. doi: 10.1039/d5sc00964b. eCollection 2025 Jun 4. Chem Sci. 2025. PMID: 40342919 Free PMC article.
-
Effective simultaneous removal of 17β-estradiol and tetracycline by a novel Alkalibacterium strain: characteristics, mechanisms, and application in livestock wastewater treatment.Appl Microbiol Biotechnol. 2025 May 27;109(1):127. doi: 10.1007/s00253-025-13516-z. Appl Microbiol Biotechnol. 2025. PMID: 40423809 Free PMC article.
-
The tetracycline resistome is shaped by selection for specific resistance mechanisms by each antibiotic generation.Nat Commun. 2025 Feb 7;16(1):1452. doi: 10.1038/s41467-025-56425-5. Nat Commun. 2025. PMID: 39920134 Free PMC article.
References
-
- Medicine, F. C. f. V. 2021 Summary Report on Antimicrobials Sold or Distributed for Use in Food-Producing Animals (U.S. Food and Drug Administration’s Center for Veterinary Medicine, 2022).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

