Maternal dietary fat during lactation shapes single nucleus transcriptomic profile of postnatal offspring hypothalamus in a sexually dimorphic manner in mice
- PMID: 38493217
- PMCID: PMC10944494
- DOI: 10.1038/s41467-024-46589-x
Maternal dietary fat during lactation shapes single nucleus transcriptomic profile of postnatal offspring hypothalamus in a sexually dimorphic manner in mice
Abstract
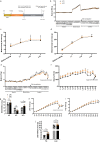
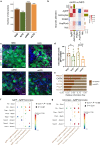
Maternal overnutrition during lactation predisposes offspring to develop metabolic diseases and exacerbates the relevant syndromes in males more than females in later life. The hypothalamus is a heterogenous brain region that regulates energy balance. Here we combined metabolic trait quantification of mother and offspring mice under low and high fat diet (HFD) feeding during lactation, with single nucleus transcriptomic profiling of their offspring hypothalamus at peak lacation to understand the cellular and molecular alterations in response to maternal dietary pertubation. We found significant expansion in neuronal subpopulations including histaminergic (Hdc), arginine vasopressin/retinoic acid receptor-related orphan receptor β (Avp/Rorb) and agouti-related peptide/neuropeptide Y (AgRP/Npy) in male offspring when their mothers were fed HFD, and increased Npy-astrocyte interactions in offspring responding to maternal overnutrition. Our study provides a comprehensive offspring hypothalamus map at the peak lactation and reveals how the cellular subpopulations respond to maternal dietary fat in a sex-specific manner during development.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- World Health Organization. Obesity and overweight. https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (2024).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

