Multifactorial genetic control and magnesium levels govern the production of a Streptomyces antibiotic with unusual cell density dependence
- PMID: 38493407
- PMCID: PMC11019849
- DOI: 10.1128/msystems.01368-23
Multifactorial genetic control and magnesium levels govern the production of a Streptomyces antibiotic with unusual cell density dependence
Abstract
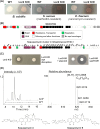
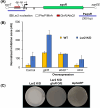
Streptomyces bacteria are renowned both for their antibiotic production capabilities and for their cryptic metabolic potential. Their metabolic repertoire is subject to stringent genetic control, with many of the associated biosynthetic gene clusters being repressed by the conserved nucleoid-associated protein Lsr2. In an effort to stimulate new antibiotic production in wild Streptomyces isolates, we leveraged the activity of an Lsr2 knockdown construct and successfully enhanced antibiotic production in the wild Streptomyces isolate WAC07094. We determined that this new activity stemmed from increased levels of the angucycline-like family member saquayamycin. Saquayamycin has both antibiotic and anti-cancer activities, and intriguingly, beyond Lsr2-mediated repression, we found saquayamycin production was also suppressed at high density on solid or in liquid growth media; its levels were greatest in low-density cultures. This density-dependent control was exerted at the level of the cluster-situated regulatory gene sqnR and was mediated in part through the activity of the PhoRP two-component regulatory system, where deleting phoRP led to both constitutive antibiotic production and sqnR expression. This suggests that PhoP functions to repress the expression of sqnR at high cell density. We further discovered that magnesium supplementation could alleviate this density dependence, although its action was independent of PhoP. Finally, we revealed that the nitrogen-responsive regulators GlnR and AfsQ1 could relieve the repression exerted by Lsr2 and PhoP. Intriguingly, we found that this low density-dependent production of saquayamycin was not unique to WAC07094; saquayamycin production by another wild isolate also exhibited low-density activation, suggesting that this spatial control may serve an important ecological function in their native environments.IMPORTANCEStreptomyces specialized metabolic gene clusters are subject to complex regulation, and their products are frequently not observed under standard laboratory growth conditions. For the wild Streptomyces isolate WAC07094, production of the angucycline-family compound saquayamycin is subject to a unique constellation of control factors. Notably, it is produced primarily at low cell density, in contrast to the high cell density production typical of most antibiotics. This unusual density dependence is conserved in other saquayamycin producers and is driven by the pathway-specific regulator SqnR, whose expression is influenced by both nutritional and genetic elements. Collectively, this work provides new insights into an intricate regulatory system governing antibiotic production and indicates there may be benefits to including low-density cultures in antibiotic screening platforms.
Keywords: Streptomyces; antibiotic; cell density; magnesium; phosphate; regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

