Loss of Dnmt3a increases self-renewal and resistance to pegIFN-α in JAK2-V617F-positive myeloproliferative neoplasms
- PMID: 38493481
- PMCID: PMC11208296
- DOI: 10.1182/blood.2023020270
Loss of Dnmt3a increases self-renewal and resistance to pegIFN-α in JAK2-V617F-positive myeloproliferative neoplasms
Abstract
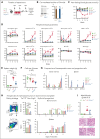
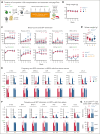
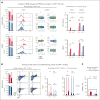
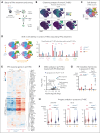
Pegylated interferon alfa (pegIFN-α) can induce molecular remissions in patients with JAK2-V617F-positive myeloproliferative neoplasms (MPNs) by targeting long-term hematopoietic stem cells (LT-HSCs). Additional somatic mutations in genes regulating LT-HSC self-renewal, such as DNMT3A, have been reported to have poorer responses to pegIFN-α. We investigated whether DNMT3A loss leads to alterations in JAK2-V617F LT-HSC functions conferring resistance to pegIFN-α treatment in a mouse model of MPN and in hematopoietic progenitors from patients with MPN. Long-term treatment with pegIFN-α normalized blood parameters and reduced splenomegaly and JAK2-V617F chimerism in single-mutant JAK2-V617F (VF) mice. However, pegIFN-α in VF;Dnmt3aΔ/Δ (VF;DmΔ/Δ) mice worsened splenomegaly and failed to reduce JAK2-V617F chimerism. Furthermore, LT-HSCs from VF;DmΔ/Δ mice compared with VF were less prone to accumulate DNA damage and exit dormancy upon pegIFN-α treatment. RNA sequencing showed that IFN-α induced stronger upregulation of inflammatory pathways in LT-HSCs from VF;DmΔ/Δ than from VF mice, indicating that the resistance of VF;DmΔ/Δ LT-HSC was not due to failure in IFN-α signaling. Transplantations of bone marrow from pegIFN-α-treated VF;DmΔ/Δ mice gave rise to more aggressive disease in secondary and tertiary recipients. Liquid cultures of hematopoietic progenitors from patients with MPN with JAK2-V617F and DNMT3A mutation showed increased percentages of JAK2-V617F-positive colonies upon IFN-α exposure, whereas in patients with JAK2-V617F alone, the percentages of JAK2-V617F-positive colonies decreased or remained unchanged. PegIFN-α combined with 5-azacytidine only partially overcame resistance in VF;DmΔ/Δ mice. However, this combination strongly decreased the JAK2-mutant allele burden in mice carrying VF mutation only, showing potential to inflict substantial damage preferentially to the JAK2-mutant clone.
© 2024 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: R.C.S. serves as a scientific adviser/scientific advisory board member and holds equity in Ajax Therapeutics; has provided consultancy services to and/or received honoraria from Novartis, Bristol Myers Squibb/Celgene, AOP, GSK, Baxalta, and Pfizer. N.H. owns stocks in the company Cantargia. The remaining authors declare no competing financial interests.
Figures




Comment in
-
DNMT3A gates IFN-induced MPN HSC exhaustion.Blood. 2024 Jun 13;143(24):2445-2446. doi: 10.1182/blood.2024024448. Blood. 2024. PMID: 38869916 No abstract available.
References
-
- Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017;129(6):667–679. - PubMed
-
- Lundberg P, Karow A, Nienhold R, et al. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123(14):2220–2228. - PubMed
-
- Guglielmelli P, Lasho TL, Rotunno G, et al. The number of prognostically detrimental mutations and prognosis in primary myelofibrosis: an international study of 797 patients. Leukemia. 2014;28(9):1804–1810. - PubMed
-
- Guglielmelli P, Lasho TL, Rotunno G, et al. MIPSS70: mutation-enhanced international prognostic score system for transplantation-age patients with primary myelofibrosis. J Clin Oncol. 2018;36(4):310–318. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

