Integrative genomic analyses identify candidate causal genes for calcific aortic valve stenosis involving tissue-specific regulation
- PMID: 38494474
- PMCID: PMC10944835
- DOI: 10.1038/s41467-024-46639-4
Integrative genomic analyses identify candidate causal genes for calcific aortic valve stenosis involving tissue-specific regulation
Abstract
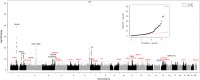
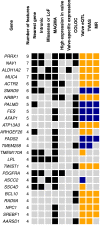
There is currently no medical therapy to prevent calcific aortic valve stenosis (CAVS). Multi-omics approaches could lead to the identification of novel molecular targets. Here, we perform a genome-wide association study (GWAS) meta-analysis including 14,819 cases among 941,863 participants of European ancestry. We report 32 genomic loci, among which 20 are novel. RNA sequencing of 500 human aortic valves highlights an enrichment in expression regulation at these loci and prioritizes candidate causal genes. Homozygous genotype for a risk variant near TWIST1, a gene involved in endothelial-mesenchymal transition, has a profound impact on aortic valve transcriptomics. We identify five genes outside of GWAS loci by combining a transcriptome-wide association study, colocalization, and Mendelian randomization analyses. Using cross-phenotype and phenome-wide approaches, we highlight the role of circulating lipoproteins, blood pressure and inflammation in the disease process. Our findings pave the way for the development of novel therapies for CAVS.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures