Genomic characterisation of the population structure and antibiotic resistance of Salmonella enterica serovar Infantis in Chile, 2009-2022
- PMID: 38495315
- PMCID: PMC10944094
- DOI: 10.1016/j.lana.2024.100711
Genomic characterisation of the population structure and antibiotic resistance of Salmonella enterica serovar Infantis in Chile, 2009-2022
Abstract
Background: Multidrug-resistant (MDR) Salmonella Infantis has disseminated worldwide, mainly linked to the consumption of poultry products. Evidence shows dissemination of this pathogen in Chile; however, studies are primarily limited to phenotypic data or involve few isolates. As human cases of Salmonella Infantis infections have substantially increased in recent years, this study aimed to characterise the genomic epidemiology and antimicrobial-resistance profiles of isolates obtained from different sources, aiming to inform effective surveillance and control measures.
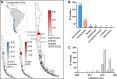
Methods: We sequenced 396 Salmonella Infantis genomes and analysed them with all publicly available genomes of this pathogen from Chile (440 genomes in total), representing isolates from environmental, food, animal, and human sources obtained from 2009 to 2022. Based on bioinformatic and phenotypic methods, we assessed the population structure, dissemination among different niches, and antimicrobial resistance (AMR) profiles of Salmonella Infantis in the country.
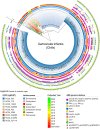
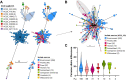
Findings: The genomic and phylogenetic analyses showed that Salmonella Infantis from Chile comprised several clusters of highly related isolates dominated by sequence type 32. The HC20_343 cluster grouped an important proportion of all isolates. This was the only cluster associated with pESI-like megaplasmids, and up to 12 acquired AMR genes/mutations predicted to result in an MDR phenotype. Accordingly, antimicrobial-susceptibility testing revealed a strong concordance between the AMR genetic determinants and their matching phenotypic expression, indicating that a significant proportion of HC20_343 isolates produce extended-spectrum β-lactamases and have intermediate fluoroquinolone resistance. HC20_343 Salmonella Infantis were spread among environmental, animal, food, and human niches, showing a close relationship between isolates from different years and sources, and a low intra-source genomic diversity.
Interpretation: Our findings show a widespread dissemination of MDR Salmonella Infantis from the HC20_343 cluster in Chile. The high proportion of isolates with resistance to first-line antibiotics and the evidence of active transmission between the environment, animals, food, and humans highlight the urgency of improved surveillance and control measures in the country. As HC20_343 isolates predominate in the Americas, our results suggest a high prevalence of ESBL-producing Salmonella Infantis with intermediate fluoroquinolone resistance in the continent.
Funding: Partially supported by the Food and Drug Administration (FDA) of the U.S. Department of Health and Human Services as part of an award, FDU001818, with 30% percent funded by FDA/HHS; and by Agencia de Investigación y Desarrollo de Chile (ANID) through FONDECYT de Postdoctorado Folio 3230796 and Folio 3210317, FONDECYT Regular Folio 1231082, and ANID-Millennium Science Initiative Program-ICN2021_044.
Keywords: Americas; Antibiotic resistance; CTX-M-65; Chile; Fluoroquinolones; Megaplasmid; Salmonella Infantis; pESI.
© 2024 The Author(s).
Conflict of interest statement
All authors declare no competing interests.
Figures

Similar articles
-
Emergence and Clonal Spread of Extended-Spectrum β-Lactamase-Producing Salmonella Infantis Carrying pESI Megaplasmids in Korean Retail Poultry Meat.Antibiotics (Basel). 2025 Apr 1;14(4):366. doi: 10.3390/antibiotics14040366. Antibiotics (Basel). 2025. PMID: 40298492 Free PMC article.
-
From farm to fork: Spread of a multidrug resistant Salmonella Infantis clone encoding blaCTX-M-1 on pESI-like plasmids in Central Italy.Int J Food Microbiol. 2024 Jan 30;410:110490. doi: 10.1016/j.ijfoodmicro.2023.110490. Epub 2023 Nov 17. Int J Food Microbiol. 2024. PMID: 37992554
-
Clonal Spread of pESI-Positive Multidrug-Resistant ST32 Salmonella enterica Serovar Infantis Isolates among Broilers and Humans in Slovenia.Microbiol Spectr. 2022 Dec 21;10(6):e0248122. doi: 10.1128/spectrum.02481-22. Epub 2022 Oct 17. Microbiol Spectr. 2022. PMID: 36250854 Free PMC article.
-
Molecular epidemiology of Salmonella Infantis in Europe: insights into the success of the bacterial host and its parasitic pESI-like megaplasmid.Microb Genom. 2020 May;6(5):e000365. doi: 10.1099/mgen.0.000365. Epub 2020 Apr 9. Microb Genom. 2020. PMID: 32271142 Free PMC article.
-
Salmonellosis: An Overview of Epidemiology, Pathogenesis, and Innovative Approaches to Mitigate the Antimicrobial Resistant Infections.Antibiotics (Basel). 2024 Jan 13;13(1):76. doi: 10.3390/antibiotics13010076. Antibiotics (Basel). 2024. PMID: 38247636 Free PMC article. Review.
Cited by
-
Salmonella in Chile: an urgent need for timely data and WGS implementation.Lancet Reg Health Am. 2025 Jul 9;48:101179. doi: 10.1016/j.lana.2025.101179. eCollection 2025 Aug. Lancet Reg Health Am. 2025. PMID: 40809294 Free PMC article. No abstract available.
-
Salmonella isolated from street foods and environment of an urban park: A whole genome sequencing approach.PLoS One. 2025 Apr 2;20(4):e0320735. doi: 10.1371/journal.pone.0320735. eCollection 2025. PLoS One. 2025. PMID: 40173163 Free PMC article.
-
Development of a robust FT-IR typing system for Salmonella enterica, enhancing performance through hierarchical classification.Microbiol Spectr. 2025 Jul;13(7):e0015925. doi: 10.1128/spectrum.00159-25. Epub 2025 May 27. Microbiol Spectr. 2025. PMID: 40422737 Free PMC article.
-
Genomic insights into antibiotic-resistant non-typhoidal Salmonella isolates from outpatients in Minhang District in Shanghai.Commun Med (Lond). 2025 Jun 12;5(1):228. doi: 10.1038/s43856-025-00950-3. Commun Med (Lond). 2025. PMID: 40506473 Free PMC article.
-
A global atlas and drivers of antimicrobial resistance in Salmonella during 1900-2023.Nat Commun. 2025 May 17;16(1):4611. doi: 10.1038/s41467-025-59758-3. Nat Commun. 2025. PMID: 40382325 Free PMC article.
References
-
- CDC . 2023. Salmonella–information for Healthcare professionals and laboratories.https://www.cdc.gov/salmonella/general/technical.html Accessed Aug 23, 2023.
-
- Aviv G., Tsyba K., Steck N., et al. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ Microbiol. 2014;16:977–994. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

