Nuclease-free precise genome editing corrects MECP2 mutations associated with Rett syndrome
- PMID: 38495533
- PMCID: PMC10940404
- DOI: 10.3389/fgeed.2024.1346781
Nuclease-free precise genome editing corrects MECP2 mutations associated with Rett syndrome
Abstract
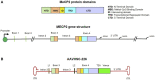
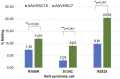
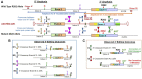
Rett syndrome is an acquired progressive neurodevelopmental disorder caused by de novo mutations in the X-linked MECP2 gene which encodes a pleiotropic protein that functions as a global transcriptional regulator and a chromatin modifier. Rett syndrome predominantly affects heterozygous females while affected male hemizygotes rarely survive. Gene therapy of Rett syndrome has proven challenging due to a requirement for stringent regulation of expression with either over- or under-expression being toxic. Ectopic expression of MECP2 in conjunction with regulatory miRNA target sequences has achieved some success, but the durability of this approach remains unknown. Here we evaluated a nuclease-free homologous recombination (HR)-based genome editing strategy to correct mutations in the MECP2 gene. The stem cell-derived AAVHSCs have previously been shown to mediate seamless and precise HR-based genome editing. We tested the ability of HR-based genome editing to correct pathogenic mutations in Exons 3 and 4 of the MECP2 gene and restore the wild type sequence while preserving all native genomic regulatory elements associated with MECP2 expression, thus potentially addressing a significant issue in gene therapy for Rett syndrome. Moreover, since the mutations are edited directly at the level of the genome, the corrections are expected to be durable with progeny cells inheriting the edited gene. The AAVHSC MECP2 editing vector was designed to be fully homologous to the target MECP2 region and to insert a promoterless Venus reporter at the end of Exon 4. Evaluation of AAVHSC editing in a panel of Rett cell lines bearing mutations in Exons 3 and 4 demonstrated successful correction and rescue of expression of the edited MECP2 gene. Sequence analysis of edited Rett cells revealed successful and accurate correction of mutations in both Exons 3 and 4 and permitted mapping of HR crossover events. Successful correction was observed only when the mutations were flanked at both the 5' and 3' ends by crossover events, but not when both crossovers occurred either exclusively upstream or downstream of the mutation. Importantly, we concluded that pathogenic mutations were successfully corrected in every Rett line analyzed, demonstrating the therapeutic potential of HR-based genome editing.
Keywords: MeCP2; Rett syndrome; adeno-associated virus; genome editing; homologous recombination.
Copyright © 2024 Bijlani, Pang, Bugga, Rangasamy, Narayanan and Chatterjee.
Conflict of interest statement
SC is an advisor to and holds equity in Homology Medicines Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
CREB Signaling Is Involved in Rett Syndrome Pathogenesis.J Neurosci. 2017 Mar 29;37(13):3671-3685. doi: 10.1523/JNEUROSCI.3735-16.2017. Epub 2017 Mar 7. J Neurosci. 2017. PMID: 28270572 Free PMC article.
-
Genetic Analysis of MECP2 Gene in Iranian Patients with Rett Syndrome.Iran J Child Neurol. 2019 Summer;13(3):25-34. Iran J Child Neurol. 2019. PMID: 31327966 Free PMC article.
-
MECP2 Mutations in the Rett Syndrome Patients from South India.Neurol India. 2022 Jan-Feb;70(1):249-253. doi: 10.4103/0028-3886.338714. Neurol India. 2022. PMID: 35263890
-
Neurodevelopmental disorders in males related to the gene causing Rett syndrome in females (MECP2).Eur J Paediatr Neurol. 2003;7(1):5-12. doi: 10.1016/s1090-3798(02)00134-4. Eur J Paediatr Neurol. 2003. PMID: 12615169 Review.
-
Associations between MeCP2 mutations, X-chromosome inactivation, and phenotype.Ment Retard Dev Disabil Res Rev. 2002;8(2):99-105. doi: 10.1002/mrdd.10026. Ment Retard Dev Disabil Res Rev. 2002. PMID: 12112735 Review.
Cited by
-
Comparative epigenetics of domestic animals: focusing on DNA accessibility and its impact on gene regulation and traits.J Vet Sci. 2025 Jan;26(1):e9. doi: 10.4142/jvs.24259. J Vet Sci. 2025. PMID: 39901471 Free PMC article. Review.
-
Site-blocking antisense oligonucleotides as a mechanism to fine-tune MeCP2 expression.RNA. 2024 Nov 18;30(12):1554-1571. doi: 10.1261/rna.080220.124. RNA. 2024. PMID: 39379106 Free PMC article.
-
Prenatal gene editing for neurodevelopmental diseases: Ethical considerations.Am J Hum Genet. 2025 Feb 6;112(2):201-214. doi: 10.1016/j.ajhg.2025.01.003. Epub 2025 Jan 28. Am J Hum Genet. 2025. PMID: 39879986 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources

