This is a preprint.
Transcription factor clusters as information transfer agents
- PMID: 38495568
- PMCID: PMC10942473
Transcription factor clusters as information transfer agents
Update in
-
Transcription factor clusters as information transfer agents.Sci Adv. 2025 Jan 3;11(1):eadp3251. doi: 10.1126/sciadv.adp3251. Epub 2025 Jan 1. Sci Adv. 2025. PMID: 39742495 Free PMC article.
Abstract
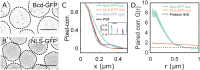
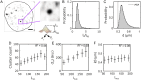
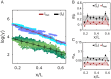
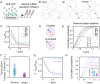
Deciphering how genes interpret information from transcription factor (TFs) concentrations within the cell nucleus remains a fundamental question in gene regulation. Recent advancements have revealed the heterogeneous distribution of TF molecules, posing challenges to precisely decoding concentration signals. Using high-resolution single-cell imaging of the fluorescently tagged TF Bicoid in living Drosophila embryos, we show that Bicoid accumulation in submicron clusters preserves the spatial information of the maternal Bicoid gradient. These clusters provide precise spatial cues through intensity, size, and frequency. We further discover that gene targets of Bicoid, such as Hunchback and Eve, colocalize with these clusters in an enhancer binding affinity-dependent manner. Our modeling suggests that clustering offers a faster sensing mechanism for global nuclear concentrations than freely diffusing TF molecules detected by simple enhancers.
Figures
References
-
- Spitz F. and Furlong E. E., Transcription factors: from enhancer binding to developmental control, Nature reviews genetics 13, 613 (2012). - PubMed
-
- Trojanowski J. and Rippe K., Transcription factor binding and activity on chromatin, Current Opinion in Systems Biology 31, 100438 (2022).
-
- Rosenfeld N., Young J. W., Alon U., Swain P. S., and Elowitz M. B., Gene Regulation at the Single-Cell Level, Science 307, 1962 (2005). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
