Construction of an aerolysin-based multi-epitope vaccine against Aeromonas hydrophila: an in silico machine learning and artificial intelligence-supported approach
- PMID: 38495891
- PMCID: PMC10940347
- DOI: 10.3389/fimmu.2024.1369890
Construction of an aerolysin-based multi-epitope vaccine against Aeromonas hydrophila: an in silico machine learning and artificial intelligence-supported approach
Abstract
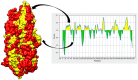
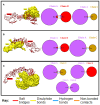
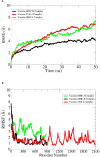
Aeromonas hydrophila, a gram-negative coccobacillus bacterium, can cause various infections in humans, including septic arthritis, diarrhea (traveler's diarrhea), gastroenteritis, skin and wound infections, meningitis, fulminating septicemia, enterocolitis, peritonitis, and endocarditis. It frequently occurs in aquatic environments and readily contacts humans, leading to high infection rates. This bacterium has exhibited resistance to numerous commercial antibiotics, and no vaccine has yet been developed. Aiming to combat the alarmingly high infection rate, this study utilizes in silico techniques to design a multi-epitope vaccine (MEV) candidate against this bacterium based on its aerolysin toxin, which is the most toxic and highly conserved virulence factor among the Aeromonas species. After retrieval, aerolysin was processed for B-cell and T-cell epitope mapping. Once filtered for toxicity, antigenicity, allergenicity, and solubility, the chosen epitopes were combined with an adjuvant and specific linkers to create a vaccine construct. These linkers and the adjuvant enhance the MEV's ability to elicit robust immune responses. Analyses of the predicted and improved vaccine structure revealed that 75.5%, 19.8%, and 1.3% of its amino acids occupy the most favored, additional allowed, and generously allowed regions, respectively, while its ERRAT score reached nearly 70%. Docking simulations showed the MEV exhibiting the highest interaction and binding energies (-1,023.4 kcal/mol, -923.2 kcal/mol, and -988.3 kcal/mol) with TLR-4, MHC-I, and MHC-II receptors. Further molecular dynamics simulations demonstrated the docked complexes' remarkable stability and maximum interactions, i.e., uniform RMSD, fluctuated RMSF, and lowest binding net energy. In silico models also predict the vaccine will stimulate a variety of immunological pathways following administration. These analyses suggest the vaccine's efficacy in inducing robust immune responses against A. hydrophila. With high solubility and no predicted allergic responses or toxicity, it appears safe for administration in both healthy and A. hydrophila-infected individuals.
Keywords: A. hydrophila; MD simulations; epitopes; immunoinformatics; systems biology; vaccine.
Copyright © 2024 Alawam and Alwethaynani.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Ahmed HA, Mohamed MEM, Rezk MM, Gharieb RMA, Abdel-Maksoud SA. Aeromonas hydrophila in fish and humans; prevalence, virulotyping and antimicrobial resistance. Slov Vet Res. (2018) 55:113–24.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

