Adaptation to seasonal reproduction and environment-associated factors drive temporal and spatial differentiation in northwest Atlantic herring despite gene flow
- PMID: 38495946
- PMCID: PMC10940790
- DOI: 10.1111/eva.13675
Adaptation to seasonal reproduction and environment-associated factors drive temporal and spatial differentiation in northwest Atlantic herring despite gene flow
Abstract
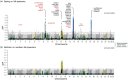
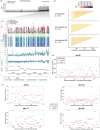
Understanding how marine organisms adapt to local environments is crucial for predicting how populations will respond to global climate change. The genomic basis, environmental factors and evolutionary processes involved in local adaptation are however not well understood. Here we use Atlantic herring, an abundant, migratory and widely distributed marine fish with substantial genomic resources, as a model organism to evaluate local adaptation. We examined genomic variation and its correlation with environmental variables across a broad environmental gradient, for 15 spawning aggregations in Atlantic Canada and the United States. We then compared our results with available genomic data of northeast Atlantic populations. We confirmed that population structure lies in a fraction of the genome including likely adaptive genetic variants of functional importance. We discovered 10 highly differentiated genomic regions distributed across four chromosomes. Nine regions show strong association with seasonal reproduction. One region, corresponding to a known inversion on chromosome 12, underlies a latitudinal pattern discriminating populations north and south of a biogeographic transition zone on the Scotian Shelf. Genome-environment associations indicate that winter seawater temperature best correlates with the latitudinal pattern of this inversion. The variation at two so-called 'islands of divergence' related to seasonal reproduction appear to be private to the northwest Atlantic. Populations in the northwest and northeast Atlantic share variation at four of these divergent regions, simultaneously displaying significant diversity in haplotype composition at another four regions, which includes an undescribed structural variant approximately 7.7 Mb long on chromosome 8. Our results suggest that the timing and geographic location of spawning and early development may be under diverse selective pressures related to allelic fitness across environments. Our study highlights the role of genomic architecture, ancestral haplotypes and selection in maintaining adaptive divergence in species with large population sizes and presumably high gene flow.
Keywords: chromosomal inversion; fisheries; genomics; marine fish; pool‐seq; whole genome.
© 2024 The Authors. Evolutionary Applications published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interest.
Figures




References
-
- Andersson, L. , Ryman, N. , Rosenberg, R. , & Ståhl, G. (1981). Genetic variability in Atlantic herring (Clupea harengus harengus): Description of protein loci and population data. Hereditas, 95(1), 69–78. 10.1111/j.1601-5223.1981.tb01330.x - DOI
-
- André, C. , Larsson, L. C. , Laikre, L. , Bekkevold, D. , Brigham, J. , Carvalho, G. R. , Dahlgren, T. G. , Hutchinson, W. F. , Mariani, S. , Mudde, K. , Ruzzante, D. E. , & Ryman, N. (2011). Detecting population structure in a high gene‐flow species, Atlantic herring (Clupea harengus): Direct, simultaneous evaluation of neutral vs putatively selected loci. Heredity, 106(2), 270–280. 10.1038/hdy.2010.71 - DOI - PMC - PubMed
-
- Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data . http://www.bioinformatics.babraham.ac.uk/projects/fastqc
-
- Bekkevold, D. , André, C. , Dahlgren, T. G. , Clausen, L. A. W. , Torstensen, E. , Mosegaard, H. , Carvalho, G. R. , Christensen, T. B. , Norlinder, E. , & Ruzzante, D. E. (2005). Environmental correlates of population differentiation in Atlantic herring. Evolution, 59(12), 2656–2668. 10.1111/j.0014-3820.2005.tb00977.x - DOI - PubMed
-
- Benestan, L. , Gosselin, T. , Perrier, C. , Sainte‐Marie, B. , Rochette, R. , & Bernatchez, L. (2015). RAD genotyping reveals fine‐scale genetic structuring and provides powerful population assignment in a widely distributed marine species, the American lobster (Homarus americanus). Molecular Ecology, 24(13), 3299–3315. 10.1111/mec.13245 - DOI - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Miscellaneous

