Why we are made of proteins and nucleic acids: Structural biology views on extraterrestrial life
- PMID: 38496239
- PMCID: PMC10941967
- DOI: 10.2142/biophysico.bppb-v20.0026
Why we are made of proteins and nucleic acids: Structural biology views on extraterrestrial life
Abstract
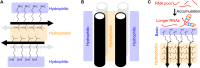
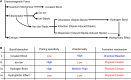
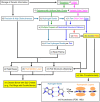
Is it a miracle that life exists on the Earth, or is it a common phenomenon in the universe? If extraterrestrial organisms exist, what are they like? To answer these questions, we must understand what kinds of molecules could evolve into life, or in other words, what properties are generally required to perform biological functions and store genetic information. This review summarizes recent findings on simple ancestral proteins, outlines the basic knowledge in textbooks, and discusses the generally required properties for biological molecules from structural biology viewpoints (e.g., restriction of shapes, and types of intra- and intermolecular interactions), leading to the conclusion that proteins and nucleic acids are at least one of the simplest (and perhaps very common) forms of catalytic and genetic biopolymers in the universe. This review article is an extended version of the Japanese article, On the Origin of Life: Coevolution between RNA and Peptide, published in SEIBUTSU BUTSURI Vol. 61, p. 232-235 (2021).
Keywords: astrobiology; origin of life.
2023 THE BIOPHYSICAL SOCIETY OF JAPAN.
Conflict of interest statement
The author declares no conflicts of interest.
Figures




Similar articles
-
Planning Implications Related to Sterilization-Sensitive Science Investigations Associated with Mars Sample Return (MSR).Astrobiology. 2022 Jun;22(S1):S112-S164. doi: 10.1089/AST.2021.0113. Epub 2022 May 19. Astrobiology. 2022. PMID: 34904892
-
Concepts of a synthetic minimal cell: Information molecules, metabolic pathways, and vesicle reproduction.Biophys Physicobiol. 2023 Dec 19;21(1):e210002. doi: 10.2142/biophysico.bppb-v21.0002. eCollection 2024. Biophys Physicobiol. 2023. PMID: 38803330 Free PMC article.
-
How evolution builds up complexity?: In vitro evolution approaches to witness complexification in artificial molecular replication systems.Biophys Physicobiol. 2022 Feb 15;19:1-10. doi: 10.2142/biophysico.bppb-v19.0005. eCollection 2022. Biophys Physicobiol. 2022. PMID: 35435608 Free PMC article.
-
Astrovirology: Viruses at Large in the Universe.Astrobiology. 2018 Feb;18(2):207-223. doi: 10.1089/ast.2017.1649. Epub 2018 Jan 10. Astrobiology. 2018. PMID: 29319335 Review.
-
Understanding prebiotic chemistry through the analysis of extraterrestrial amino acids and nucleobases in meteorites.Chem Soc Rev. 2012 Aug 21;41(16):5459-72. doi: 10.1039/c2cs35109a. Epub 2012 Jun 15. Chem Soc Rev. 2012. PMID: 22706603 Review.
Cited by
-
An ancestral fold reveals the evolutionary link between RNA polymerase and ribosomal proteins.Nat Commun. 2024 Jul 18;15(1):5938. doi: 10.1038/s41467-024-50013-9. Nat Commun. 2024. PMID: 39025855 Free PMC article.
References
-
- Greenwald, J., Kwiatkowski, W., Riek, R.. Peptide amyloids in the origin of life. J. Mol. Biol. 430, 3735–3750 (2018). https://doi.org/10.1016/j.jmb.2018.05.046 - PubMed
-
- Despotovic, D., Tawfik, D. S.. Proto‐proteins in protocells. ChemSystemsChem 3, e2100002 (2021). https://doi.org/10.1002/syst.202100002
-
- Tagami, S., Li, P.. The origin of life: RNA and protein co‐evolution on the ancient earth. Dev. Growth Differ. 65, 167–174 (2023). https://doi.org/10.1111/dgd.12845 - PubMed
-
- Brack, A., Orgel, L. E.. β structures of alternating polypeptides and their possible prebiotic significance. Nature 256, 383–387 (1975). https://doi.org/10.1038/256383a0 - PubMed
-
- DeGrado, W. F., Wasserman, Z. R., Lear, J. D.. Protein design, a minimalist approach. Science 243, 622–628 (1989). https://doi.org/10.1126/science.2464850 - PubMed

