This is a preprint.
Lipid Peroxidation and Type I Interferon Coupling Fuels Pathogenic Macrophage Activation Causing Tuberculosis Susceptibility
- PMID: 38496444
- PMCID: PMC10942339
- DOI: 10.1101/2024.03.05.583602
Lipid Peroxidation and Type I Interferon Coupling Fuels Pathogenic Macrophage Activation Causing Tuberculosis Susceptibility
Abstract
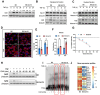
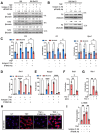
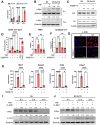
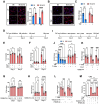
A quarter of human population is infected with Mycobacterium tuberculosis, but less than 10% of those infected develop pulmonary TB. We developed a genetically defined sst1-susceptible mouse model that uniquely reproduces a defining feature of human TB: the development of necrotic lung granulomas and determined that the sst1-susceptible phenotype was driven by the aberrant macrophage activation. This study demonstrates that the aberrant response of the sst1-susceptible macrophages to prolonged stimulation with TNF is primarily driven by conflicting Myc and antioxidant response pathways leading to a coordinated failure 1) to properly sequester intracellular iron and 2) to activate ferroptosis inhibitor enzymes. Consequently, iron-mediated lipid peroxidation fueled Ifnβ superinduction and sustained the Type I Interferon (IFN-I) pathway hyperactivity that locked the sst1-susceptible macrophages in a state of unresolving stress and compromised their resistance to Mtb. The accumulation of the aberrantly activated, stressed, macrophages within granuloma microenvironment led to the local failure of anti-tuberculosis immunity and tissue necrosis. The upregulation of Myc pathway in peripheral blood cells of human TB patients was significantly associated with poor outcomes of TB treatment. Thus, Myc dysregulation in activated macrophages results in an aberrant macrophage activation and represents a novel target for host-directed TB therapies.
Keywords: Myc; antioxidant defense; inflammation; lipid peroxidation; macrophage; tuberculosis; type I interferon.
Conflict of interest statement
Declaration of interests: The authors have declared that no conflict of interest exists.
Figures


Similar articles
-
TLR2 is non-redundant in the population and subpopulation responses to Mycobacterium tuberculosis in macrophages and in vivo.mSystems. 2023 Aug 31;8(4):e0005223. doi: 10.1128/msystems.00052-23. Epub 2023 Jul 13. mSystems. 2023. PMID: 37439558 Free PMC article.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Concurrent TB and HIV therapies effectively control clinical reactivation of TB during co-infection but fail to eliminate chronic immune activation.Res Sq [Preprint]. 2024 Aug 26:rs.3.rs-4908400. doi: 10.21203/rs.3.rs-4908400/v1. Res Sq. 2024. PMID: 39257997 Free PMC article. Preprint.
-
Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults.Cochrane Database Syst Rev. 2014 Jan 21;2014(1):CD009593. doi: 10.1002/14651858.CD009593.pub3. Cochrane Database Syst Rev. 2014. Update in: Cochrane Database Syst Rev. 2019 Jun 07;6:CD009593. doi: 10.1002/14651858.CD009593.pub4. PMID: 24448973 Free PMC article. Updated.
-
Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults.Cochrane Database Syst Rev. 2013 Jan 31;(1):CD009593. doi: 10.1002/14651858.CD009593.pub2. Cochrane Database Syst Rev. 2013. Update in: Cochrane Database Syst Rev. 2014 Jan 21;(1):CD009593. doi: 10.1002/14651858.CD009593.pub3. PMID: 23440842 Free PMC article. Updated.
References
-
- Alic N, Partridge L (2015) Myc mouse and anti-ageing therapy. Trends in Endocrinology & Metabolism 26: 163 – 164 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
