This is a preprint.
Comparison of Synergy Extrapolation and Static Optimization for Estimating Multiple Unmeasured Muscle Activations during Walking
- PMID: 38496460
- PMCID: PMC10942366
- DOI: 10.1101/2024.03.03.583228
Comparison of Synergy Extrapolation and Static Optimization for Estimating Multiple Unmeasured Muscle Activations during Walking
Update in
-
Comparison of synergy extrapolation and static optimization for estimating multiple unmeasured muscle activations during walking.J Neuroeng Rehabil. 2024 Nov 1;21(1):194. doi: 10.1186/s12984-024-01490-y. J Neuroeng Rehabil. 2024. PMID: 39482723 Free PMC article.
Abstract
Background: Calibrated electromyography (EMG)-driven musculoskeletal models can provide great insight into internal quantities (e.g., muscle forces) that are difficult or impossible to measure experimentally. However, the need for EMG data from all involved muscles presents a significant barrier to the widespread application of EMG-driven modeling methods. Synergy extrapolation (SynX) is a computational method that can estimate a single missing EMG signal with reasonable accuracy during the EMG-driven model calibration process, yet its performance in estimating a larger number of missing EMG signals remains unclear.
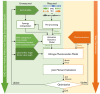
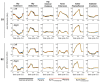
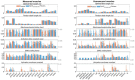
Methods: This study assessed the accuracy with which SynX can use eight measured EMG signals to estimate muscle activations and forces associated with eight missing EMG signals in the same leg during walking while simultaneously performing EMG-driven model calibration. Experimental gait data collected from two individuals post-stroke, including 16 channels of EMG data per leg, were used to calibrate an EMG-driven musculoskeletal model, providing "gold standard" muscle activations and forces for evaluation purposes. SynX was then used to predict the muscle activations and forces associated with the eight missing EMG signals while simultaneously calibrating EMG-driven model parameter values. Due to its widespread use, static optimization (SO) was also utilized to estimate the same muscle activations and forces. Estimation accuracy for SynX and SO was evaluated using root mean square errors (RMSE) to quantify amplitude errors and correlation coefficient values to quantify shape similarity, each calculated with respect to "gold standard" muscle activations and forces.
Results: On average, SynX produced significantly more accurate amplitude and shape estimates for unmeasured muscle activations (RMSE 0.08 vs. value 0.55 vs. 0.12) and forces (RMSE 101.3 N vs. value 0.53 vs. 0.07) compared to SO. SynX yielded calibrated Hill-type muscle-tendon model parameter values for all muscles and activation dynamics model parameter values for measured muscles that were similar to "gold standard" calibrated model parameter values.
Conclusions: These findings suggest that SynX could make it possible to calibrate EMG-driven musculoskeletal models for all important lower-extremity muscles with as few as eight carefully chosen EMG signals and eventually contribute to the design of personalized rehabilitation and surgical interventions for mobility impairments.
Keywords: EMG-driven model; Model personalization; Muscle activation; Muscle force; Static optimization; Stroke; Synergy extrapolation.
Conflict of interest statement
Competing interests The authors declare that they have no competing interests.
Figures






Similar articles
-
Comparison of synergy extrapolation and static optimization for estimating multiple unmeasured muscle activations during walking.J Neuroeng Rehabil. 2024 Nov 1;21(1):194. doi: 10.1186/s12984-024-01490-y. J Neuroeng Rehabil. 2024. PMID: 39482723 Free PMC article.
-
EMG-driven musculoskeletal model calibration with estimation of unmeasured muscle excitations via synergy extrapolation.Front Bioeng Biotechnol. 2022 Sep 7;10:962959. doi: 10.3389/fbioe.2022.962959. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36159690 Free PMC article.
-
Evaluation of Synergy Extrapolation for Predicting Unmeasured Muscle Excitations from Measured Muscle Synergies.Front Comput Neurosci. 2020 Dec 4;14:588943. doi: 10.3389/fncom.2020.588943. eCollection 2020. Front Comput Neurosci. 2020. PMID: 33343322 Free PMC article.
-
A computational method for estimating trunk muscle activations during gait using lower extremity muscle synergies.Front Bioeng Biotechnol. 2022 Dec 13;10:964359. doi: 10.3389/fbioe.2022.964359. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 36582837 Free PMC article.
-
Contributions to the understanding of gait control.Dan Med J. 2014 Apr;61(4):B4823. Dan Med J. 2014. PMID: 24814597 Review.
References
-
- Pandy MG, Lin Y-C, Kim HJ. Muscle coordination of mediolateral balance in normal walking. J Biomech. 2010;43:2055–64. - PubMed
-
- Kim HJ, Fernandez JW, Akbarshahi M, Walter JP, Fregly BJ, Pandy MG. Evaluation of predicted knee-joint muscle forces during gait using an instrumented knee implant. J Orthop Res. 2009;27:1326–31. - PubMed
-
- Marra MA, Vanheule V, Fluit R, Koopman BH, Rasmussen J, Verdonschot N, et al. A subject-specific musculoskeletal modeling framework to predict in vivo mechanics of total knee arthroplasty. J Biomech Eng. 2015;137:020904. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
