This is a preprint.
Assessment of the evidence yield for the calibrated PP3/BP4 computational recommendations
- PMID: 38496501
- PMCID: PMC10942508
- DOI: 10.1101/2024.03.05.24303807
Assessment of the evidence yield for the calibrated PP3/BP4 computational recommendations
Update in
-
Assessment of the evidence yield for the calibrated PP3/BP4 computational recommendations.Genet Med. 2024 Nov;26(11):101213. doi: 10.1016/j.gim.2024.101213. Epub 2024 Jul 25. Genet Med. 2024. PMID: 39030733
Abstract
Purpose: To investigate the number of rare missense variants observed in human genome sequences by ACMG/AMP PP3/BP4 evidence strength, following the calibrated PP3/BP4 computational recommendations.
Methods: Missense variants from the genome sequences of 300 probands from the Rare Genomes Project with suspected rare disease were analyzed using computational prediction tools able to reach PP3_Strong and BP4_Moderate evidence strengths (BayesDel, MutPred2, REVEL, and VEST4). The numbers of variants at each evidence strength were analyzed across disease-associated genes and genome-wide.
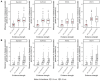
Results: From a median of 75.5 rare (≤1% allele frequency) missense variants in disease-associated genes per proband, a median of one reached PP3_Strong, 3-5 PP3_Moderate, and 3-5 PP3_Supporting. Most were allocated BP4 evidence (median 41-49 per proband) or were indeterminate (median 17.5-19 per proband). Extending the analysis to all protein-coding genes genome-wide, the number of PP3_Strong variants increased approximately 2.6-fold compared to disease-associated genes, with a median per proband of 1-3 PP3_Strong, 8-16 PP3_Moderate, and 10-17 PP3_Supporting.
Conclusion: A small number of variants per proband reached PP3_Strong and PP3_Moderate in 3,424 disease-associated genes, and though not the intended use of the recommendations, also genome-wide. Use of PP3/BP4 evidence as recommended from calibrated computational prediction tools in the clinical diagnostic laboratory is unlikely to inappropriately contribute to the classification of an excessive number of variants as Pathogenic or Likely Pathogenic by ACMG/AMP rules.
Conflict of interest statement
CONFLICT OF INTEREST Disclosure: L.G.B. receives royalties from Wolters-Kluwer for authorship of UpToDate, is a member of the Illumina Medical Ethics Committee, and receives research support from Merck, Inc. V.P. and P.R. participated in the development of some of the tools assessed in this study. All other authors declare no conflict of interest.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
