This is a preprint.
Histology-guided mathematical model of tumor oxygenation: sensitivity analysis of physical and computational parameters
- PMID: 38496532
- PMCID: PMC10942376
- DOI: 10.1101/2024.03.05.583363
Histology-guided mathematical model of tumor oxygenation: sensitivity analysis of physical and computational parameters
Abstract
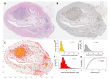
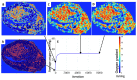
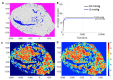
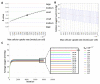
A hybrid off-lattice agent-based model has been developed to reconstruct the tumor tissue oxygenation landscape based on histology images and simulated interactions between vasculature and cells with microenvironment metabolites. Here, we performed a robustness sensitivity analysis of that model's physical and computational parameters. We found that changes in the domain boundary conditions, the initial conditions, and the Michaelis constant are negligible and, thus, do not affect the model outputs. The model is also not sensitive to small perturbations of the vascular influx or the maximum consumption rate of oxygen. However, the model is sensitive to large perturbations of these parameters and changes in the tissue boundary condition, emphasizing an imperative aim to measure these parameters experimentally.
Figures



References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
