This is a preprint.
Targeting LINC00152 activates cAMP/Ca2+/ferroptosis axis and overcomes tamoxifen resistance in ER+ breast cancer
- PMID: 38496603
- PMCID: PMC10942410
- DOI: 10.1101/2023.11.05.565697
Targeting LINC00152 activates cAMP/Ca2+/ferroptosis axis and overcomes tamoxifen resistance in ER+ breast cancer
Update in
-
Targeting LINC00152 activates cAMP/Ca2+/ferroptosis axis and overcomes tamoxifen resistance in ER+ breast cancer.Cell Death Dis. 2024 Jun 15;15(6):418. doi: 10.1038/s41419-024-06814-3. Cell Death Dis. 2024. PMID: 38879508 Free PMC article.
Abstract
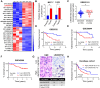
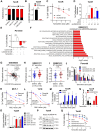
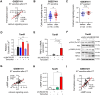
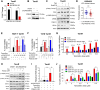
Tamoxifen has been the mainstay therapy to treat early, locally advanced, and metastatic estrogen receptor-positive (ER+) breast cancer, constituting around 75% of all cases. However, emergence of resistance is common, necessitating the identification of novel therapeutic targets. Here, we demonstrated that long-noncoding RNA LINC00152 confers tamoxifen resistance via blocking tamoxifen-induced ferroptosis, an iron-mediated cell death. Mechanistically, inhibiting LINC00152 reduces the mRNA stability of phosphodiesterase 4D (PDE4D), leading to activation of cAMP/PKA/CREB axis and increased expression of TRPC1 Ca2+ channel. This causes cytosolic Ca2+ overload and generation of reactive oxygen species (ROS) that is, on one hand, accompanied by downregulation of FTH1, a member of the iron sequestration unit, thus increasing intracellular Fe2+ levels; and on the other hand, inhibition of the peroxidase activity upon reduced GPX4 and xCT levels. These ultimately induce lipid peroxidation and ferroptotic cell death in combination with tamoxifen. Overexpressing PDE4D rescues LINC00152 inhibition-mediated tamoxifen sensitization by de-activating the cAMP/Ca2+/ferroptosis axis. Importantly, high LINC00152 expression is significantly correlated with high PDE4D/low ferroptosis and worse survival in multiple cohorts of tamoxifen- or tamoxifen-containing endocrine therapy-treated ER+ breast cancer patients. Overall, we identified LINC00152 inhibition as a novel mechanism of ferroptosis induction and tamoxifen sensitization, thereby revealing LINC00152 and its effectors as actionable therapeutic targets to improve clinical outcome in refractory ER+ breast cancer.
Keywords: ER+ breast cancer; cAMP; calcium; ferroptosis; lncRNA; tamoxifen resistance.
Conflict of interest statement
Competing interests: O.S. is the co-founder of OncoCube Therapeutics LLC, the founder and president of LoxiGen, Inc and the scientific advisory board member of A2A Pharmaceuticals. The other authors declare no potential conflict of interest.
Figures


References
-
- Clemons M., Danson S. & Howell A. Tamoxifen (“Nolvadex”): a review. Cancer treatment reviews 28, 165–180 (2002). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
