This is a preprint.
Targeted, Genome-scale Overexpression in Proteobacteria
- PMID: 38496613
- PMCID: PMC10942329
- DOI: 10.1101/2024.03.01.582922
Targeted, Genome-scale Overexpression in Proteobacteria
Abstract
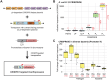
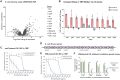
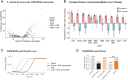
Targeted, genome-scale gene perturbation screens using Clustered Regularly Interspaced Short Palindromic Repeats interference (CRISPRi) and activation (CRISPRa) have revolutionized eukaryotic genetics, advancing medical, industrial, and basic research. Although CRISPRi knockdowns have been broadly applied in bacteria, options for genome-scale gene overexpression face key limitations. Here, we develop a facile approach for genome-scale overexpression in bacteria we call, "CRISPRtOE" (CRISPR transposition and OverExpression). We first create a platform for comprehensive gene targeting using CRISPR-associated transposons (CAST) and show that transposition occurs at a higher frequency in non-transcribed DNA. We then demonstrate that CRISPRtOE can upregulate gene expression in Proteobacteria with medical and industrial relevance by integrating synthetic promoters of varying strength upstream of target genes. Finally, we employ CRISPRtOE screening at the genome-scale in the model bacterium Escherichia coli and the non-model biofuel producer Zymomonas mobilis, recovering known and novel antibiotic and engineering targets. We envision that CRISPRtOE will be a valuable overexpression tool for antibiotic mode of action, industrial strain optimization, and gene function discovery in bacteria.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures




References
-
- Jost M, Chen Y, Gilbert LA, Horlbeck MA, Krenning L, Menchon G, Rai A, Cho MY, Stern JJ, Prota AE, Kampmann M, Akhmanova A, Steinmetz MO, Tanenbaum ME, Weissman JS. 2017. Combined CRISPRi/a-Based Chemical Genetic Screens Reveal that Rigosertib Is a Microtubule-Destabilizing Agent. Molecular Cell 68:210–223.e6. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
