This is a preprint.
Ongoing evolution of SARS-CoV-2 drives escape from mRNA vaccine-induced humoral immunity
- PMID: 38496628
- PMCID: PMC10942518
- DOI: 10.1101/2024.03.05.24303815
Ongoing evolution of SARS-CoV-2 drives escape from mRNA vaccine-induced humoral immunity
Update in
-
Ongoing evolution of SARS-CoV-2 drives escape from mRNA vaccine-induced humoral immunity.Cell Rep Med. 2024 Dec 17;5(12):101850. doi: 10.1016/j.xcrm.2024.101850. Epub 2024 Dec 9. Cell Rep Med. 2024. PMID: 39657661 Free PMC article.
Abstract
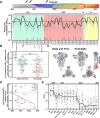
Since the COVID-19 pandemic began in 2020, viral sequencing has documented 131 individual mutations in the viral spike protein across 48 named variants. To determine the ability of vaccine-mediated humoral immunity to keep pace with continued SARS-CoV-2 evolution, we assessed the neutralization potency of sera from 76 vaccine recipients collected after 2 to 6 immunizations against a comprehensive panel of mutations observed during the pandemic. Remarkably, while many individual mutations that emerged between 2020 and 2022 exhibit escape from sera following primary vaccination, few escape boosted sera. However, progressive loss of neutralization was observed across newer variants, irrespective of vaccine doses. Importantly, an updated XBB.1.5 booster significantly increased titers against newer variants but not JN.1. These findings demonstrate that seasonal boosters improve titers against contemporaneous strains, but novel variants continue to evade updated mRNA vaccines, demonstrating the need for novel approaches to adequately control SARS-CoV-2 transmission.
Keywords: COVID-19; JN.1; SARS-CoV-2; XBB.1.5 Booster; breadth; infectivity; neutralizing antibodies; spike; vaccination; variants.
Conflict of interest statement
DECLARATIONS OF INTEREST A.B.B. is a founder of Cure Systems LLC.
Figures





Similar articles
-
Ongoing evolution of SARS-CoV-2 drives escape from mRNA vaccine-induced humoral immunity.Cell Rep Med. 2024 Dec 17;5(12):101850. doi: 10.1016/j.xcrm.2024.101850. Epub 2024 Dec 9. Cell Rep Med. 2024. PMID: 39657661 Free PMC article.
-
Human and hamster sera correlate well in identifying antigenic drift among SARS-CoV-2 variants, including JN.1.J Virol. 2024 Nov 19;98(11):e0094824. doi: 10.1128/jvi.00948-24. Epub 2024 Oct 4. J Virol. 2024. PMID: 39365051 Free PMC article.
-
mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant.Cell. 2022 Feb 3;185(3):457-466.e4. doi: 10.1016/j.cell.2021.12.033. Epub 2022 Jan 6. Cell. 2022. PMID: 34995482 Free PMC article.
-
Diversified humoral immunity and impacts of booster vaccines: SARS-CoV-2 antibody profile and Omicron BA.2 neutralization before and after first or second boosters.Microbiol Spectr. 2024 Oct 3;12(10):e0060524. doi: 10.1128/spectrum.00605-24. Epub 2024 Aug 20. Microbiol Spectr. 2024. PMID: 39162540 Free PMC article.
-
Neutralizing antibody responses and cellular responses against SARS-CoV-2 Omicron subvariants after mRNA SARS-CoV-2 vaccination in kidney transplant recipients.Sci Rep. 2024 May 28;14(1):12176. doi: 10.1038/s41598-024-63147-z. Sci Rep. 2024. PMID: 38806644 Free PMC article.
References
-
- WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int.
-
- Worldometers.info (2023). Worldometer - FAQ. https://www.worldometers.info/faq/.
-
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. (2020). Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 182, 812–827.e19. 10.1016/j.cell.2020.06.043. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
