This is a preprint.
Cryo-EM confirms a common fibril fold in the heart of four patients with ATTRwt amyloidosis
- PMID: 38496656
- PMCID: PMC10942412
- DOI: 10.1101/2024.03.08.582936
Cryo-EM confirms a common fibril fold in the heart of four patients with ATTRwt amyloidosis
Update in
-
Cryo-EM confirms a common fibril fold in the heart of four patients with ATTRwt amyloidosis.Commun Biol. 2024 Jul 27;7(1):905. doi: 10.1038/s42003-024-06588-6. Commun Biol. 2024. PMID: 39068302 Free PMC article.
Abstract
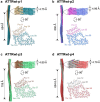
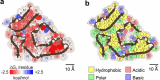
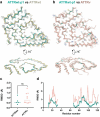
ATTR amyloidosis results from the conversion of transthyretin into amyloid fibrils that deposit in tissues causing organ failure and death. This conversion is facilitated by mutations in ATTRv amyloidosis, or aging in ATTRwt amyloidosis. ATTRv amyloidosis exhibits extreme phenotypic variability, whereas ATTRwt amyloidosis presentation is consistent and predictable. Previously, we found an unprecedented structural variability in cardiac amyloid fibrils from polyneuropathic ATTRv-I84S patients. In contrast, cardiac fibrils from five genotypically-different patients with cardiomyopathy or mixed phenotypes are structurally homogeneous. To understand fibril structure's impact on phenotype, it is necessary to study the fibrils from multiple patients sharing genotype and phenotype. Here we show the cryo-electron microscopy structures of fibrils extracted from four cardiomyopathic ATTRwt amyloidosis patients. Our study confirms that they share identical conformations with minimal structural variability, consistent with their homogenous clinical presentation. Our study contributes to the understanding of ATTR amyloidosis biopathology and calls for further studies.
Figures


References
-
- González-López E., et al. Wild-type transthyretin amyloidosis as a cause of heart failure with preserved ejection fraction. European Heart Journal 36, 2585–2594 (2015). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
