Roles for epithelial integrin α3β1 in regulation of the microenvironment during normal and pathological tissue remodeling
- PMID: 38497112
- PMCID: PMC11371326
- DOI: 10.1152/ajpcell.00128.2024
Roles for epithelial integrin α3β1 in regulation of the microenvironment during normal and pathological tissue remodeling
Abstract
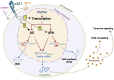
Integrin receptors for the extracellular matrix activate intracellular signaling pathways that are critical for tissue development, homeostasis, and regeneration/repair, and their loss or dysregulation contributes to many developmental defects and tissue pathologies. This review will focus on tissue remodeling roles for integrin α3β1, a receptor for laminins found in the basement membranes (BMs) that underlie epithelial cell layers. As a paradigm, we will discuss literature that supports a role for α3β1 in promoting ability of epidermal keratinocytes to modify their tissue microenvironment during skin development, wound healing, or tumorigenesis. Preclinical and clinical studies have shown that this role depends largely on ability of α3β1 to govern the keratinocyte's repertoire of secreted proteins, or the "secretome," including 1) matrix proteins and proteases involved in matrix remodeling and 2) paracrine-acting growth factors/cytokines that stimulate other cells with important tissue remodeling functions (e.g., endothelial cells, fibroblasts, inflammatory cells). Moreover, α3β1 signaling controls gene expression that helps epithelial cells carry out these functions, including genes that encode secreted matrix proteins, proteases, growth factors, or cytokines. We will review what is known about α3β1-dependent gene regulation through both transcription and posttranscriptional mRNA stability. Regarding the latter, we will discuss examples of α3β1-dependent alternative splicing (AS) or alternative polyadenylation (APA) that prevents inclusion of cis-acting mRNA sequences that would otherwise target the transcript for degradation via nonsense-mediated decay or destabilizing AU-rich elements (AREs) in the 3'-untranslated region (3'-UTR). Finally, we will discuss prospects and anticipated challenges of exploiting α3β1 as a clinical target for the treatment of cancer or wound healing.
Keywords: alternative polyadenylation; alternative splicing; integrin α3β1; secretome; tissue microenvironment.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures

Similar articles
-
Integrin α3β1 signaling through MEK/ERK determines alternative polyadenylation of the MMP-9 mRNA transcript in immortalized mouse keratinocytes.PLoS One. 2015 Mar 9;10(3):e0119539. doi: 10.1371/journal.pone.0119539. eCollection 2015. PLoS One. 2015. PMID: 25751421 Free PMC article.
-
The epidermal integrin-mediated secretome regulates the skin microenvironment during tumorigenesis and repair.Matrix Biol. 2024 Dec;134:175-183. doi: 10.1016/j.matbio.2024.11.002. Epub 2024 Nov 2. Matrix Biol. 2024. PMID: 39491760 Review.
-
Alpha3beta1 integrin in epidermis promotes wound angiogenesis and keratinocyte-to-endothelial-cell crosstalk through the induction of MRP3.J Cell Sci. 2009 Jun 1;122(Pt 11):1778-87. doi: 10.1242/jcs.040956. Epub 2009 May 12. J Cell Sci. 2009. PMID: 19435806 Free PMC article.
-
Integrin alpha3beta1 potentiates TGFbeta-mediated induction of MMP-9 in immortalized keratinocytes.J Invest Dermatol. 2008 Mar;128(3):575-86. doi: 10.1038/sj.jid.5701042. Epub 2007 Aug 30. J Invest Dermatol. 2008. PMID: 17762853 Free PMC article.
-
Integrin-mediated regulation of epidermal wound functions.Cell Tissue Res. 2016 Sep;365(3):467-82. doi: 10.1007/s00441-016-2446-2. Epub 2016 Jun 28. Cell Tissue Res. 2016. PMID: 27351421 Free PMC article. Review.
Cited by
-
Engineering cell-derived extracellular matrix for peripheral nerve regeneration.Mater Today Bio. 2024 Jun 13;27:101125. doi: 10.1016/j.mtbio.2024.101125. eCollection 2024 Aug. Mater Today Bio. 2024. PMID: 38979129 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

