Application of MALDI-TOF MS and machine learning for the detection of SARS-CoV-2 and non-SARS-CoV-2 respiratory infections
- PMID: 38497716
- PMCID: PMC11064577
- DOI: 10.1128/spectrum.04068-23
Application of MALDI-TOF MS and machine learning for the detection of SARS-CoV-2 and non-SARS-CoV-2 respiratory infections
Abstract
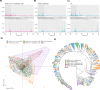
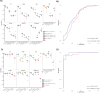
Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) could aid the diagnosis of acute respiratory infections (ARIs) owing to its affordability and high-throughput capacity. MALDI-TOF MS has been proposed for use on commonly available respiratory samples, without specialized sample preparation, making this technology especially attractive for implementation in low-resource regions. Here, we assessed the utility of MALDI-TOF MS in differentiating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vs non-COVID acute respiratory infections (NCARIs) in a clinical lab setting in Kazakhstan. Nasopharyngeal swabs were collected from inpatients and outpatients with respiratory symptoms and from asymptomatic controls (ACs) in 2020-2022. PCR was used to differentiate SARS-CoV-2+ and NCARI cases. MALDI-TOF MS spectra were obtained for a total of 252 samples (115 SARS-CoV-2+, 98 NCARIs, and 39 ACs) without specialized sample preparation. In our first sub-analysis, we followed a published protocol for peak preprocessing and machine learning (ML), trained on publicly available spectra from South American SARS-CoV-2+ and NCARI samples. In our second sub-analysis, we trained ML models on a peak intensity matrix representative of both South American (SA) and Kazakhstan (Kaz) samples. Applying the established MALDI-TOF MS pipeline "as is" resulted in a high detection rate for SARS-CoV-2+ samples (91.0%), but low accuracy for NCARIs (48.0%) and ACs (67.0%) by the top-performing random forest model. After re-training of the ML algorithms on the SA-Kaz peak intensity matrix, the accuracy of detection by the top-performing support vector machine with radial basis function kernel model was at 88.0%, 95.0%, and 78% for the Kazakhstan SARS-CoV-2+, NCARI, and AC subjects, respectively, with a SARS-CoV-2 vs rest receiver operating characteristic area under the curve of 0.983 [0.958, 0.987]; a high differentiation accuracy was maintained for the South American SARS-CoV-2 and NCARIs. MALDI-TOF MS/ML is a feasible approach for the differentiation of ARI without specialized sample preparation. The implementation of MALDI-TOF MS/ML in a real clinical lab setting will necessitate continuous optimization to keep up with the rapidly evolving landscape of ARI.IMPORTANCEIn this proof-of-concept study, the authors used matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) and machine learning (ML) to identify and distinguish acute respiratory infections (ARI) caused by SARS-CoV-2 versus other pathogens in low-resource clinical settings, without the need for specialized sample preparation. The ML models were trained on a varied collection of MALDI-TOF MS spectra from studies conducted in Kazakhstan and South America. Initially, the MALDI-TOF MS/ML pipeline, trained exclusively on South American samples, exhibited diminished effectiveness in recognizing non-SARS-CoV-2 infections from Kazakhstan. Incorporation of spectral signatures from Kazakhstan substantially increased the accuracy of detection. These results underscore the potential of employing MALDI-TOF MS/ML in resource-constrained settings to augment current approaches for detecting and differentiating ARI.
Keywords: COVID-19; MALDI-TOF MS; SARS-CoV-2; acute respiratory infection; machine learning.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Novel application of automated machine learning with MALDI-TOF-MS for rapid high-throughput screening of COVID-19: a proof of concept.Sci Rep. 2021 Apr 15;11(1):8219. doi: 10.1038/s41598-021-87463-w. Sci Rep. 2021. PMID: 33859233 Free PMC article.
-
A combined approach of MALDI-TOF mass spectrometry and multivariate analysis as a potential tool for the detection of SARS-CoV-2 virus in nasopharyngeal swabs.J Virol Methods. 2020 Dec;286:113991. doi: 10.1016/j.jviromet.2020.113991. Epub 2020 Oct 9. J Virol Methods. 2020. PMID: 33045283 Free PMC article.
-
Detection of SARS-CoV-2 in nasal swabs using MALDI-MS.Nat Biotechnol. 2020 Oct;38(10):1168-1173. doi: 10.1038/s41587-020-0644-7. Epub 2020 Jul 30. Nat Biotechnol. 2020. PMID: 32733106
-
Use of MALDI-TOF mass spectrometry for virus identification: a review.Analyst. 2022 Jul 12;147(14):3131-3154. doi: 10.1039/d2an00431c. Analyst. 2022. PMID: 35713185 Review.
-
Machine learning for microbial identification and antimicrobial susceptibility testing on MALDI-TOF mass spectra: a systematic review.Clin Microbiol Infect. 2020 Oct;26(10):1310-1317. doi: 10.1016/j.cmi.2020.03.014. Epub 2020 Mar 23. Clin Microbiol Infect. 2020. PMID: 32217160
Cited by
-
Complete genome sequence of a human influenza a virus (H1N1) detected in Kazakhstan in the fall of 2022.Microbiol Resour Announc. 2025 Jan 16;14(1):e0104024. doi: 10.1128/mra.01040-24. Epub 2024 Dec 10. Microbiol Resour Announc. 2025. PMID: 39655925 Free PMC article.
-
Metagenomic profiling of nasopharyngeal samples from adults with acute respiratory infection.R Soc Open Sci. 2024 Jul 10;11(7):240108. doi: 10.1098/rsos.240108. eCollection 2024 Jul. R Soc Open Sci. 2024. PMID: 39076360 Free PMC article.
-
Innovations in MALDI-TOF Mass Spectrometry: Bridging modern diagnostics and historical insights.Open Life Sci. 2025 Jul 18;20(1):20251136. doi: 10.1515/biol-2025-1136. eCollection 2025. Open Life Sci. 2025. PMID: 40688408 Free PMC article. Review.
References
-
- Yegorov S, Goremykina M, Ivanova R, Good SV, Babenko D, Shevtsov A, MacDonald KS, Zhunussov Y, COVID-19 Genomics and Semey COVID-19 Epidemiology Research Groups . 2021. Epidemiology, clinical characteristics, and virologic features of COVID-19 patients in Kazakhstan: a nation-wide retrospective cohort study. Lancet Reg Health Eur 4:100096. doi:10.1016/j.lanepe.2021.100096 - DOI - PMC - PubMed
-
- Hanson KE, Azar MM, Banerjee R, Chou A, Colgrove RC, Ginocchio CC, Hayden MK, Holodiny M, Jain S, Koo S, Levy J, Timbrook TT, Caliendo AM. 2020. Molecular testing for acute respiratory tract infections: clinical and diagnostic recommendations from the IDSA’s diagnostics committee. Clin Infect Dis 71:2744–2751. doi:10.1093/cid/ciaa508 - DOI - PMC - PubMed
-
- Deulofeu M, García-Cuesta E, Peña-Méndez EM, Conde JE, Jiménez-Romero O, Verdú E, Serrando MT, Salvadó V, Boadas-Vaello P. 2021. Detection of SARS-CoV-2 infection in human nasopharyngeal samples by combining MALDI-TOF MS and artificial intelligence. Front Med 8:661358. doi:10.3389/fmed.2021.661358 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

