The phospholipids cardiolipin and phosphatidylethanolamine differentially regulate MDC biogenesis
- PMID: 38497895
- PMCID: PMC10949074
- DOI: 10.1083/jcb.202302069
The phospholipids cardiolipin and phosphatidylethanolamine differentially regulate MDC biogenesis
Abstract
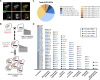
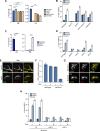
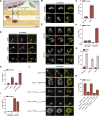
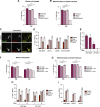
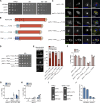
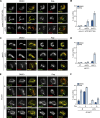
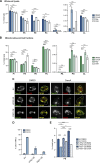
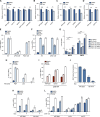
Cells utilize multiple mechanisms to maintain mitochondrial homeostasis. We recently characterized a pathway that remodels mitochondria in response to metabolic alterations and protein overload stress. This remodeling occurs via the formation of large membranous structures from the mitochondrial outer membrane called mitochondrial-derived compartments (MDCs), which are eventually released from mitochondria and degraded. Here, we conducted a microscopy-based screen in budding yeast to identify factors that regulate MDC formation. We found that two phospholipids, cardiolipin (CL) and phosphatidylethanolamine (PE), differentially regulate MDC biogenesis. CL depletion impairs MDC biogenesis, whereas blocking mitochondrial PE production leads to constitutive MDC formation. Additionally, in response to metabolic MDC activators, cellular and mitochondrial PE declines, and overexpressing mitochondrial PE synthesis enzymes suppress MDC biogenesis. Altogether, our data indicate a requirement for CL in MDC biogenesis and suggest that PE depletion may stimulate MDC formation downstream of MDC-inducing metabolic stress.
© 2024 Xiao et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures