A Top-Down Proteomic Assay to Evaluate KRAS4B-Compound Engagement
- PMID: 38498381
- PMCID: PMC10993199
- DOI: 10.1021/acs.analchem.3c05626
A Top-Down Proteomic Assay to Evaluate KRAS4B-Compound Engagement
Abstract
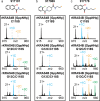
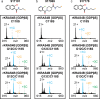
Development of new targeted inhibitors for oncogenic KRAS mutants may benefit from insight into how a given mutation influences the accessibility of protein residues and how compounds interact with mutant or wild-type KRAS proteins. Targeted proteomic analysis, a key validation step in the KRAS inhibitor development process, typically involves both intact mass- and peptide-based methods to confirm compound localization or quantify binding. However, these methods may not always provide a clear picture of the compound binding affinity for KRAS, how specific the compound is to the target KRAS residue, and how experimental conditions may impact these factors. To address this, we have developed a novel top-down proteomic assay to evaluate in vitro KRAS4B-compound engagement while assessing relative quantitation in parallel. We present two applications to demonstrate the capabilities of our assay: maleimide-biotin labeling of a KRAS4BG12D cysteine mutant panel and treatment of three KRAS4B proteins (WT, G12C, and G13C) with small molecule compounds. Our results show the time- or concentration-dependence of KRAS4B-compound engagement in context of the intact protein molecule while directly mapping the compound binding site.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

