Transposon sequencing reveals metabolic pathways essential for Mycobacterium tuberculosis infection
- PMID: 38498580
- PMCID: PMC10977890
- DOI: 10.1371/journal.ppat.1011663
Transposon sequencing reveals metabolic pathways essential for Mycobacterium tuberculosis infection
Abstract
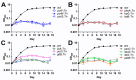
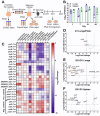
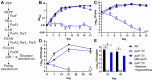
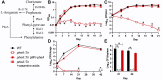
New drugs are needed to shorten and simplify treatment of tuberculosis caused by Mycobacterium tuberculosis. Metabolic pathways that M. tuberculosis requires for growth or survival during infection represent potential targets for anti-tubercular drug development. Genes and metabolic pathways essential for M. tuberculosis growth in standard laboratory culture conditions have been defined by genome-wide genetic screens. However, whether M. tuberculosis requires these essential genes during infection has not been comprehensively explored because mutant strains cannot be generated using standard methods. Here we show that M. tuberculosis requires the phenylalanine (Phe) and de novo purine and thiamine biosynthetic pathways for mammalian infection. We used a defined collection of M. tuberculosis transposon (Tn) mutants in essential genes, which we generated using a custom nutrient-rich medium, and transposon sequencing (Tn-seq) to identify multiple central metabolic pathways required for fitness in a mouse infection model. We confirmed by individual retesting and complementation that mutations in pheA (Phe biosynthesis) or purF (purine and thiamine biosynthesis) cause death of M. tuberculosis in the absence of nutrient supplementation in vitro and strong attenuation in infected mice. Our findings show that Tn-seq with defined Tn mutant pools can be used to identify M. tuberculosis genes required during mouse lung infection. Our results also demonstrate that M. tuberculosis requires Phe and purine/thiamine biosynthesis for survival in the host, implicating these metabolic pathways as prime targets for the development of new antibiotics to combat tuberculosis.
Copyright: © 2024 Block et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

References
-
- WHO. Global Tuberculosis Report 2023. Geneva: World Health Organization; 2023.
-
- WHO. WHO consolidated guidlines on tuberculosis. Module 4: treatment—drug-susceptible tuberculosis treatment. Geneva: World Health Organization; 2022. - PubMed
-
- WHO. WHO consolidated guidelines on tuberculosis. Module 4: treatment—drug-resistant tuberculosis treatment, 2022 update. Geneva: World Health Organization; 2022. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

