Sleep deprivation drives brain-wide changes in cholinergic presynapse abundance in Drosophila melanogaster
- PMID: 38498719
- PMCID: PMC10990117
- DOI: 10.1073/pnas.2312664121
Sleep deprivation drives brain-wide changes in cholinergic presynapse abundance in Drosophila melanogaster
Abstract
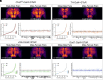
Sleep is an evolutionarily conserved state that supports brain functions, including synaptic plasticity, in species across the animal kingdom. Here, we examine the neuroanatomical and cell-type distribution of presynaptic scaling in the fly brain after sleep loss. We previously found that sleep loss drives accumulation of the active zone scaffolding protein Bruchpilot (BRP) within cholinergic Kenyon cells of the Drosophila melanogaster mushroom body (MB), but not in other classes of MB neurons. To test whether similar cell type-specific trends in plasticity occur broadly across the brain, we used a flp-based genetic reporter to label presynaptic BRP in cholinergic, dopaminergic, GABAergic, or glutamatergic neurons. We then collected whole-brain confocal image stacks of BRP intensity to systematically quantify BRP, a marker of presynapse abundance, across 37 neuropil regions of the central fly brain. Our results indicate that sleep loss, either by overnight (12-h) mechanical stimulation or chronic sleep disruption in insomniac mutants, broadly elevates cholinergic synapse abundance across the brain, while synapse abundance in neurons that produce other neurotransmitters undergoes weaker, if any, changes. Extending sleep deprivation to 24 h drives brain-wide upscaling in glutamatergic, but not other, synapses. Finally, overnight male-male social pairings induce increased BRP in excitatory synapses despite male-female pairings eliciting more waking activity, suggesting experience-specific plasticity. Within neurotransmitter class and waking context, BRP changes are similar across the 37 neuropil domains, indicating that similar synaptic scaling rules may apply across the brain during acute sleep loss and that sleep need may broadly alter excitatory-inhibitory balance in the central brain.
Keywords: Drosophila; plasticity; sleep.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
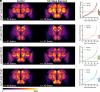
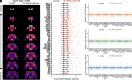
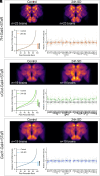

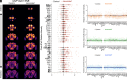
Similar articles
-
Sleep deprivation results in diverse patterns of synaptic scaling across the Drosophila mushroom bodies.Curr Biol. 2021 Aug 9;31(15):3248-3261.e3. doi: 10.1016/j.cub.2021.05.018. Epub 2021 Jun 8. Curr Biol. 2021. PMID: 34107302 Free PMC article.
-
CRY-dependent plasticity of tetrad presynaptic sites in the visual system of Drosophila at the morning peak of activity and sleep.Sci Rep. 2020 Oct 23;10(1):18161. doi: 10.1038/s41598-020-74442-w. Sci Rep. 2020. PMID: 33097794 Free PMC article.
-
Presynaptic Active Zone Plasticity Encodes Sleep Need in Drosophila.Curr Biol. 2020 Mar 23;30(6):1077-1091.e5. doi: 10.1016/j.cub.2020.01.019. Epub 2020 Mar 5. Curr Biol. 2020. PMID: 32142702
-
Synaptic organization of the mushroom body calyx in Drosophila melanogaster.J Comp Neurol. 2002 Apr 8;445(3):211-26. doi: 10.1002/cne.10155. J Comp Neurol. 2002. PMID: 11920702
-
Synapse development and maturation at the drosophila neuromuscular junction.Neural Dev. 2020 Aug 2;15(1):11. doi: 10.1186/s13064-020-00147-5. Neural Dev. 2020. PMID: 32741370 Free PMC article. Review.
Cited by
-
Developing forebrain synapses are uniquely vulnerable to sleep loss.Proc Natl Acad Sci U S A. 2024 Oct 29;121(44):e2407533121. doi: 10.1073/pnas.2407533121. Epub 2024 Oct 23. Proc Natl Acad Sci U S A. 2024. PMID: 39441640 Free PMC article.
-
miR218-5p tips the scales.Proc Natl Acad Sci U S A. 2025 May 13;122(19):e2506039122. doi: 10.1073/pnas.2506039122. Epub 2025 May 5. Proc Natl Acad Sci U S A. 2025. PMID: 40324096 No abstract available.
-
Coupling of mitochondrial state with active zone plasticity in early brain aging.Redox Biol. 2025 Feb;79:103454. doi: 10.1016/j.redox.2024.103454. Epub 2024 Dec 3. Redox Biol. 2025. PMID: 39642596 Free PMC article.
-
Socialization causes long-lasting behavioral changes.Sci Rep. 2024 Sep 27;14(1):22302. doi: 10.1038/s41598-024-73218-w. Sci Rep. 2024. PMID: 39333212 Free PMC article.
-
Alcohol induces long-lasting sleep deficits in Drosophila via subsets of cholinergic neurons.Curr Biol. 2025 Mar 10;35(5):1033-1046.e3. doi: 10.1016/j.cub.2025.01.026. Epub 2025 Feb 6. Curr Biol. 2025. PMID: 39919743
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

