Truncation of the constant domain drives amyloid formation by immunoglobulin light chains
- PMID: 38499153
- PMCID: PMC11016911
- DOI: 10.1016/j.jbc.2024.107174
Truncation of the constant domain drives amyloid formation by immunoglobulin light chains
Abstract
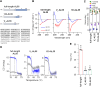
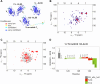
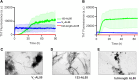
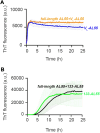
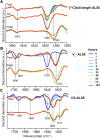
AL amyloidosis is a life-threatening disease caused by deposition of immunoglobulin light chains. While the mechanisms underlying light chains amyloidogenesis in vivo remain unclear, several studies have highlighted the role that tissue environment and structural amyloidogenicity of individual light chains have in the disease pathogenesis. AL natural deposits contain both full-length light chains and fragments encompassing the variable domain (VL) as well as different length segments of the constant region (CL), thus highlighting the relevance that proteolysis may have in the fibrillogenesis pathway. Here, we investigate the role of major truncated species of the disease-associated AL55 light chain that were previously identified in natural deposits. Specifically, we study structure, molecular dynamics, thermal stability, and capacity to form fibrils of a fragment containing both the VL and part of the CL (133-AL55), in comparison with the full-length protein and its variable domain alone, under shear stress and physiological conditions. Whereas the full-length light chain forms exclusively amorphous aggregates, both fragments generate fibrils, although, with different kinetics, aggregate structure, and interplay with the unfragmented protein. More specifically, the VL-CL 133-AL55 fragment entirely converts into amyloid fibrils microscopically and spectroscopically similar to their ex vivo counterpart and increases the amorphous aggregation of full-length AL55. Overall, our data support the idea that light chain structure and proteolysis are both relevant for amyloidogenesis in vivo and provide a novel biocompatible model of light chain fibrillogenesis suitable for future mechanistic studies.
Keywords: AL amyloidosis; amyloidogenesis; cardiomyopathy; constant domain; immunoglobulin light chains; proteolysis.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Buxbaum J.N., Dispenzieri A., Eisenberg D.S., Fändrich M., Merlini G., Saraiva M.J.M., et al. Amyloid nomenclature 2022: update, novel proteins, and recommendations by the international society of amyloidosis (ISA) nomenclature committee. Amyloid. 2022;29:213–219. - PubMed
-
- Merlini G., Dispenzieri A., Sanchorawala V., Schönland S.O., Palladini G., Hawkins P.N., et al. Systemic immunoglobulin light chain amyloidosis. Nat. Rev. Dis. Primers. 2018;4:38. - PubMed
-
- Bellotti V., Mangione P., Merlini G. Review: immunoglobulin light chain amyloidosis--the archetype of structural and pathogenic variability. J. Struct. Biol. 2000;130:280–289. - PubMed
-
- Kourelis T.V., Dasari S., Theis J.D., Ramirez-Alvarado M., Kurtin P.J., Gertz M.A., et al. Clarifying immunoglobulin gene usage in systemic and localized immunoglobulin light-chain amyloidosis by mass spectrometry. Blood. 2017;129:299–306. - PubMed
-
- Kyle R.A., Greipp P.R. “Idiopathic” Bence Jones proteinuria: long-term follow-up in seven patients. N. Engl. J. Med. 1982;306:564–567. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

