Biomimetic Platelet-Cloaked Nanoparticles for the Delivery of Anti-Inflammatory Curcumin in the Treatment of Atherosclerosis
- PMID: 38499190
- PMCID: PMC11468963
- DOI: 10.1002/adhm.202302074
Biomimetic Platelet-Cloaked Nanoparticles for the Delivery of Anti-Inflammatory Curcumin in the Treatment of Atherosclerosis
Abstract
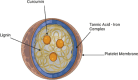
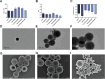
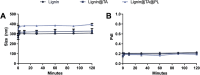
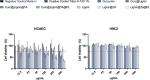
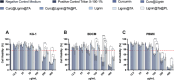
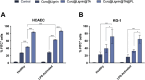
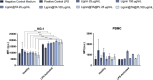
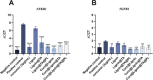
Atherosclerosis still represents a major driver of cardiovascular diseases worldwide. Together with accumulation of lipids in the plaque, inflammation is recognized as one of the key players in the formation and development of atherosclerotic plaque. Systemic anti-inflammatory treatments are successful in reducing the disease burden, but are correlated with severe side effects, underlining the need for targeted formulations. In this work, curcumin is chosen as the anti-inflammatory payload model and further loaded in lignin-based nanoparticles (NPs). The NPs are then coated with a tannic acid (TA)- Fe (III) complex and further cloaked with fragments derived from platelet cell membrane, yielding NPs with homogenous size. The two coatings increase the interaction between the NPs and cells, both endothelial and macrophages, in steady state or inflamed status. Furthermore, NPs are cytocompatible toward endothelial, smooth muscle and immune cells, while not inducing immune activation. The anti-inflammatory efficacy is demonstrated in endothelial cells by real-time quantitative polymerase chain reaction and ELISA assay where curcumin-loaded NPs decrease the expression of Nf-κb, TGF-β1, IL-6, and IL-1β in lipopolysaccharide-inflamed cells. Overall, due to the increase in the cell-NP interactions and the anti-inflammatory efficacy, these NPs represent potential candidates for the targeted anti-inflammatory treatment of atherosclerosis.
Keywords: anti‐inflammatory; atherosclerosis; biohybrid; nanoparticles; platelets membrane.
© 2024 The Authors. Advanced Healthcare Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

