A genetic screen to uncover mechanisms underlying lipid transfer protein function at membrane contact sites
- PMID: 38499328
- PMCID: PMC10948934
- DOI: 10.26508/lsa.202302525
A genetic screen to uncover mechanisms underlying lipid transfer protein function at membrane contact sites
Abstract
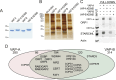
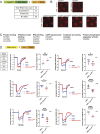
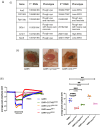
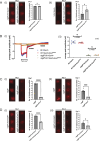
Lipid transfer proteins mediate the transfer of lipids between organelle membranes, and the loss of function of these proteins has been linked to neurodegeneration. However, the mechanism by which loss of lipid transfer activity leads to neurodegeneration is not understood. In Drosophila photoreceptors, depletion of retinal degeneration B (RDGB), a phosphatidylinositol transfer protein, leads to defective phototransduction and retinal degeneration, but the mechanism by which loss of this activity leads to retinal degeneration is not understood. RDGB is localized to membrane contact sites through the interaction of its FFAT motif with the ER integral protein VAP. To identify regulators of RDGB function in vivo, we depleted more than 300 VAP-interacting proteins and identified a set of 52 suppressors of rdgB The molecular identity of these suppressors indicates a role of novel lipids in regulating RDGB function and of transcriptional and ubiquitination processes in mediating retinal degeneration in rdgB9 The human homologs of several of these molecules have been implicated in neurodevelopmental diseases underscoring the importance of VAP-mediated processes in these disorders.
© 2024 Mishra et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Cabukusta B, Berlin I, van Elsland DM, Forkink I, Spits M, de Jong AWM, Akkermans JJLL, Wijdeven RHM, Janssen GMC, van Veelen PA, et al. (2020) Human VAPome analysis reveals MOSPD1 and MOSPD3 as membrane contact site proteins interacting with FFAT-related FFNT motifs. Cell Rep 33: 108475. 10.1016/j.celrep.2020.108475 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
