Decoding human in vitro terminal erythropoiesis originating from umbilical cord blood mononuclear cells and pluripotent stem cells
- PMID: 38499435
- PMCID: PMC11216933
- DOI: 10.1111/cpr.13614
Decoding human in vitro terminal erythropoiesis originating from umbilical cord blood mononuclear cells and pluripotent stem cells
Abstract
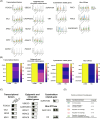
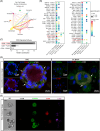
Ex vivo red blood cell (RBC) production generates unsatisfactory erythroid cells. A deep exploration into terminally differentiated cells is required to understand the impairments for RBC generation and the underlying mechanisms. Here, we mapped an atlas of terminally differentiated cells from umbilical cord blood mononuclear cells (UCBMN) and pluripotent stem cells (PSC) and observed their dynamic regulation of erythropoiesis at single-cell resolution. Interestingly, we detected a few progenitor cells and non-erythroid cells from both origins. In PSC-derived erythropoiesis (PSCE), the expression of haemoglobin switch regulators (BCL11A and ZBTB7A) were significantly absent, which could be the restraint for its adult globin expression. We also found that PSCE were less active in stress erythropoiesis than in UCBMN-derived erythropoiesis (UCBE), and explored an agonist of stress erythropoiesis gene, TRIB3, could enhance the expression of adult globin in PSCE. Compared with UCBE, there was a lower expression of epigenetic-related proteins (e.g., CASPASE 3 and UBE2O) and transcription factors (e.g., FOXO3 and TAL1) in PSCE, which might restrict PSCE's enucleation. Moreover, we characterized a subpopulation with high proliferation capacity marked by CD99high in colony-forming unit-erythroid cells. Inhibition of CD99 reduced the proliferation of PSC-derived cells and facilitated erythroid maturation. Furthermore, CD99-CD99 mediated the interaction between macrophages and erythroid cells, illustrating a mechanism by which macrophages participate in erythropoiesis. This study provided a reference for improving ex vivo RBC generation.
© 2024 The Authors. Cell Proliferation published by Beijing Institute for Stem Cell and Regenerative Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




References
-
- Rallapalli S, Guhathakurta S, Narayan S, Bishi DK, Balasubramanian V, Korrapati PS. Generation of clinical‐grade red blood cells from human umbilical cord blood mononuclear cells. Cell Tissue Res. 2019;375(2):437‐449. - PubMed
-
- Jacobsen SEW, Nerlov C. Haematopoiesis in the era of advanced single‐cell technologies. Nat Cell Biol. 2019;21(1):2‐8. - PubMed
MeSH terms
Grants and funding
- 202002030025/Science and Technology Program of Guangzhou, China
- 2017YFA0103100/National Key Research and Development Program of China
- 2017YFA0103103/National Key Research and Development Program of China
- 2017YFA0103104/National Key Research and Development Program of China
- 2022YFC2406803/National Key Research and Development Program of China
- XDA16010602/Strategic Priority Research Program of the Chinese Academy of Sciences
- 81870097/National Natural Science Foundation of China
- 82070114/National Natural Science Foundation of China
- 82270126/National Natural Science Foundation of China
- 32200589/National Natural Science Foundation of China
- 82200690/National Natural Science Foundation of China
- 32300612/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Research Materials

