Y-box binding protein 1 in small extracellular vesicles reduces mesenchymal stem cell differentiation to osteoblasts-implications for acute myeloid leukaemia
- PMID: 38499475
- PMCID: PMC10948369
- DOI: 10.1002/jev2.12417
Y-box binding protein 1 in small extracellular vesicles reduces mesenchymal stem cell differentiation to osteoblasts-implications for acute myeloid leukaemia
Abstract
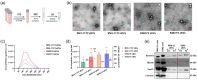
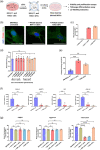
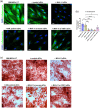
Small extracellular vesicles (sEVs) released by acute myeloid leukaemia (AML) cells have been reported to influence the trilineage differentiation of bone marrow-derived mesenchymal stem cells (BM-MSCs). However, it remains elusive which biological cargo from AML-sEVs is responsible for this effect. In this study, sEVs were isolated from cell-conditioned media and blood plasma using size-exclusion chromatography and ultrafiltration and characterized according to MISEV2018 guidelines. Our results demonstrated that AML-sEVs increased the proliferation of BM-MSCs. Conversely, key proteins that are important for normal haematopoiesis were downregulated in BM-MSCs. Additionally, we revealed that AML-sEVs significantly reduced the differentiation of BM-MSCs to osteoblasts without affecting adipogenic or chondrogenic differentiation. Next, LC-MS/MS proteomics elucidated that various proteins, including Y-box-binding protein 1 (YBX1), were upregulated in both AML-sEVs and BM-MSCs treated with AML-sEVs. Clinically relevant, we found that YBX1 is considerably upregulated in most paediatric AML patient-derived sEVs compared to healthy controls. Interestingly, sEVs isolated after the downregulation of YBX1 in AML cells remarkably rescued the osteoblastic differentiation of BM-MSCs. Altogether, our data demonstrate for the first time that YBX1 containing AML-sEVs is one of the key players that disrupt the normal function of bone marrow microenvironment by reducing the osteogenic differentiation of BM-MSCs.
Keywords: Y-box binding protein 1; acute myeloid leukaemia; bone marrow microenvironment; mesenchymal stem cells; osteoblasts; small extracellular vesicles.
© 2024 The Authors. Journal of Extracellular Vesicles published by Wiley Periodicals LLC on behalf of International Society for Extracellular Vesicles.
Conflict of interest statement
The authors report no conflict of interest.
Figures


References
-
- Antony‐Debre, I. , Bluteau, D. , Itzykson, R. , Baccini, V. , Renneville, A. , Boehlen, F. , Morabito, M. , Droin, N. , Deswarte, C. , Chang, Y. , Leverger, G. , Solary, E. , Vainchenker, W. , Favier, R. , & Raslova, H. (2012). MYH10 protein expression in platelets as a biomarker of RUNX1 and FLI1 alterations. Blood, 120(13), 2719–2722. - PubMed
-
- Azevedo, P. L. , Dias, R. B. , Nogueira, L. P. , Maradei, S. , Bigni, R. , Aragao, J. S. , Abdelhay, E. , & Binato, R. (2022). Reduced osteogenic differentiation potential in vivo in acute myeloid leukaemia patients correlates with decreased BMP4 expression in mesenchymal stromal cells. International Journal of Stem Cells, 15(2), 227–232. - PMC - PubMed
-
- BioRender . Available from: https://app.biorender.com/
-
- Bluteau, D. , Glembotsky, A. C. , Raimbault, A. , Balayn, N. , Gilles, L. , Rameau, P. , Nurden, P. , Alessi, M. C. , Debili, N. , Vainchenker, W. , Heller, P. G. , Favier, R. , & Raslova, H. (2012). Dysmegakaryopoiesis of FPD/AML pedigrees with constitutional RUNX1 mutations is linked to myosin II deregulated expression. Blood, 120(13), 2708–2718. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

