Multiscale topology in interactomic network: from transcriptome to antiaddiction drug repurposing
- PMID: 38499497
- PMCID: PMC10948341
- DOI: 10.1093/bib/bbae054
Multiscale topology in interactomic network: from transcriptome to antiaddiction drug repurposing
Abstract
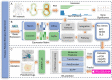
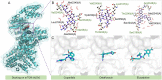
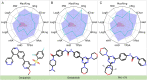
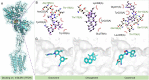
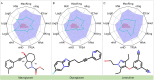
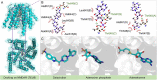
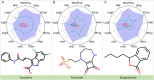
The escalating drug addiction crisis in the United States underscores the urgent need for innovative therapeutic strategies. This study embarked on an innovative and rigorous strategy to unearth potential drug repurposing candidates for opioid and cocaine addiction treatment, bridging the gap between transcriptomic data analysis and drug discovery. We initiated our approach by conducting differential gene expression analysis on addiction-related transcriptomic data to identify key genes. We propose a novel topological differentiation to identify key genes from a protein-protein interaction network derived from DEGs. This method utilizes persistent Laplacians to accurately single out pivotal nodes within the network, conducting this analysis in a multiscale manner to ensure high reliability. Through rigorous literature validation, pathway analysis and data-availability scrutiny, we identified three pivotal molecular targets, mTOR, mGluR5 and NMDAR, for drug repurposing from DrugBank. We crafted machine learning models employing two natural language processing (NLP)-based embeddings and a traditional 2D fingerprint, which demonstrated robust predictive ability in gauging binding affinities of DrugBank compounds to selected targets. Furthermore, we elucidated the interactions of promising drugs with the targets and evaluated their drug-likeness. This study delineates a multi-faceted and comprehensive analytical framework, amalgamating bioinformatics, topological data analysis and machine learning, for drug repurposing in addiction treatment, setting the stage for subsequent experimental validation. The versatility of the methods we developed allows for applications across a range of diseases and transcriptomic datasets.
Keywords: differentially expressed gene; drug repurposing; persistent spectral theory; substance addiction.
© The Author(s) 2024. Published by Oxford University Press.
Figures




Similar articles
-
Machine-learning repurposing of DrugBank compounds for opioid use disorder.Comput Biol Med. 2023 Jun;160:106921. doi: 10.1016/j.compbiomed.2023.106921. Epub 2023 May 2. Comput Biol Med. 2023. PMID: 37178605 Free PMC article.
-
Using Drug Expression Profiles and Machine Learning Approach for Drug Repurposing.Methods Mol Biol. 2019;1903:219-237. doi: 10.1007/978-1-4939-8955-3_13. Methods Mol Biol. 2019. PMID: 30547445 Review.
-
TIDAL: Topology-Inferred Drug Addiction Learning.J Chem Inf Model. 2023 Mar 13;63(5):1472-1489. doi: 10.1021/acs.jcim.3c00046. Epub 2023 Feb 24. J Chem Inf Model. 2023. PMID: 36826415
-
Unraveling new avenues in pancreatic cancer treatment: A comprehensive exploration of drug repurposing using transcriptomic data.Comput Biol Med. 2025 Feb;185:109481. doi: 10.1016/j.compbiomed.2024.109481. Epub 2024 Dec 6. Comput Biol Med. 2025. PMID: 39644581
-
[Data-driven drug discovery for drug repurposing].Nihon Yakurigaku Zasshi. 2023;158(1):10-14. doi: 10.1254/fpj.22072. Nihon Yakurigaku Zasshi. 2023. PMID: 36596476 Review. Japanese.
Cited by
-
Artificial intelligence approaches for anti-addiction drug discovery.Digit Discov. 2025 May 13;4(6):1404-1416. doi: 10.1039/d5dd00032g. eCollection 2025 Jun 11. Digit Discov. 2025. PMID: 40401266 Free PMC article. Review.
-
Hodge Decomposition of Single-Cell RNA Velocity.J Chem Inf Model. 2024 Apr 22;64(8):3558-3568. doi: 10.1021/acs.jcim.4c00132. Epub 2024 Apr 4. J Chem Inf Model. 2024. PMID: 38572676 Free PMC article.
-
Proteomic Learning of Gamma-Aminobutyric Acid (GABA) Receptor-Mediated Anesthesia.J Chem Inf Model. 2025 Apr 14;65(7):3655-3668. doi: 10.1021/acs.jcim.5c00114. Epub 2025 Mar 17. J Chem Inf Model. 2025. PMID: 40094320 Free PMC article.
References
-
- McLellan AT, Lewis DC, O’Brien CP, Kleber HD. Drug dependence, a chronic medical illness: implications for treatment, insurance, and outcomes evaluation. JAMA 2000;284(13):1689–95. - PubMed
-
- Zomorodian A, Carlsson G. Computing persistent homology. Discrete Comput. Geom, 2005;33(2):249–274.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

