Expanding the flexibility of base editing for high-throughput genetic screens in bacteria
- PMID: 38499498
- PMCID: PMC11039988
- DOI: 10.1093/nar/gkae174
Expanding the flexibility of base editing for high-throughput genetic screens in bacteria
Abstract
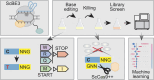
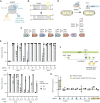
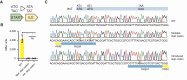
Genome-wide screens have become powerful tools for elucidating genotype-to-phenotype relationships in bacteria. Of the varying techniques to achieve knockout and knockdown, CRISPR base editors are emerging as promising options. However, the limited number of available, efficient target sites hampers their use for high-throughput screening. Here, we make multiple advances to enable flexible base editing as part of high-throughput genetic screening in bacteria. We first co-opt the Streptococcus canis Cas9 that exhibits more flexible protospacer-adjacent motif recognition than the traditional Streptococcus pyogenes Cas9. We then expand beyond introducing premature stop codons by mutating start codons. Next, we derive guide design rules by applying machine learning to an essentiality screen conducted in Escherichia coli. Finally, we rescue poorly edited sites by combining base editing with Cas9-induced cleavage of unedited cells, thereby enriching for intended edits. The efficiency of this dual system was validated through a conditional essentiality screen based on growth in minimal media. Overall, expanding the scope of genome-wide knockout screens with base editors could further facilitate the investigation of new gene functions and interactions in bacteria.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
A recombineering-based platform for high-throughput genomic editing in Escherichia coli.Appl Environ Microbiol. 2025 Jul 23;91(7):e0019325. doi: 10.1128/aem.00193-25. Epub 2025 Jun 12. Appl Environ Microbiol. 2025. PMID: 40503884 Free PMC article.
-
Rapid two-step target capture ensures efficient CRISPR-Cas9-guided genome editing.Mol Cell. 2025 May 1;85(9):1730-1742.e9. doi: 10.1016/j.molcel.2025.03.024. Epub 2025 Apr 23. Mol Cell. 2025. PMID: 40273916 Free PMC article.
-
Orthogonal transcriptional modulation and gene editing using multiple CRISPR-Cas systems.Mol Ther. 2025 Jan 8;33(1):71-89. doi: 10.1016/j.ymthe.2024.11.024. Epub 2024 Nov 19. Mol Ther. 2025. PMID: 39563029
-
Application of CHyMErA Cas9-Cas12a combinatorial genome-editing platform for genetic interaction mapping and gene fragment deletion screening.Nat Protoc. 2021 Oct;16(10):4722-4765. doi: 10.1038/s41596-021-00595-1. Epub 2021 Sep 10. Nat Protoc. 2021. PMID: 34508260 Free PMC article. Review.
-
Artificial Intelligence in CRISPR-Cas Systems: A Review of Tool Applications.Methods Mol Biol. 2025;2952:243-257. doi: 10.1007/978-1-0716-4690-8_14. Methods Mol Biol. 2025. PMID: 40553337 Review.
Cited by
-
The rise and future of CRISPR-based approaches for high-throughput genomics.FEMS Microbiol Rev. 2024 Sep 18;48(5):fuae020. doi: 10.1093/femsre/fuae020. FEMS Microbiol Rev. 2024. PMID: 39085047 Free PMC article. Review.
References
-
- Przybyla L., Gilbert L.A. A new era in functional genomics screens. Nat. Rev. Genet. 2022; 23:89–103. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

