Will we ever be able to accurately predict solubility?
- PMID: 38499581
- PMCID: PMC10948805
- DOI: 10.1038/s41597-024-03105-6
Will we ever be able to accurately predict solubility?
Abstract
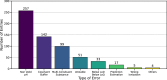
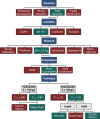
Accurate prediction of thermodynamic solubility by machine learning remains a challenge. Recent models often display good performances, but their reliability may be deceiving when used prospectively. This study investigates the origins of these discrepancies, following three directions: a historical perspective, an analysis of the aqueous solubility dataverse and data quality. We investigated over 20 years of published solubility datasets and models, highlighting overlooked datasets and the overlaps between popular sets. We benchmarked recently published models on a novel curated solubility dataset and report poor performances. We also propose a workflow to cure aqueous solubility data aiming at producing useful models for bench chemist. Our results demonstrate that some state-of-the-art models are not ready for public usage because they lack a well-defined applicability domain and overlook historical data sources. We report the impact of factors influencing the utility of the models: interlaboratory standard deviation, ionic state of the solute and data sources. The herein obtained models, and quality-assessed datasets are publicly available.
© 2024. The Author(s).
Conflict of interest statement
C. Minoletti and P. Llompart are Sanofi employees and may hold shares and/or stock options in the company. S. Baybekov, D. Horvath, G. Marcou, and A. Varnek have nothing to disclose.
Figures

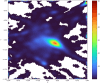






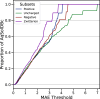


References
-
- Kennedy T. Managing the drug discovery/development interface. Drug Discov. Today. 1997;2:436–444. doi: 10.1016/S1359-6446(97)01099-4. - DOI
-
- Jouyban A, Abolghassemi Fakhree MA. Solubility prediction methods for drug/drug like molecules. Recent Pat. Chem. Eng. 2008;1:220–231. doi: 10.2174/2211334710801030220. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

