Ferritin heavy chain supports stability and function of the regulatory T cell lineage
- PMID: 38499786
- PMCID: PMC11021483
- DOI: 10.1038/s44318-024-00064-x
Ferritin heavy chain supports stability and function of the regulatory T cell lineage
Abstract
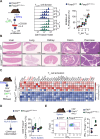
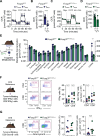
Regulatory T (TREG) cells develop via a program orchestrated by the transcription factor forkhead box protein P3 (FOXP3). Maintenance of the TREG cell lineage relies on sustained FOXP3 transcription via a mechanism involving demethylation of cytosine-phosphate-guanine (CpG)-rich elements at conserved non-coding sequences (CNS) in the FOXP3 locus. This cytosine demethylation is catalyzed by the ten-eleven translocation (TET) family of dioxygenases, and it involves a redox reaction that uses iron (Fe) as an essential cofactor. Here, we establish that human and mouse TREG cells express Fe-regulatory genes, including that encoding ferritin heavy chain (FTH), at relatively high levels compared to conventional T helper cells. We show that FTH expression in TREG cells is essential for immune homeostasis. Mechanistically, FTH supports TET-catalyzed demethylation of CpG-rich sequences CNS1 and 2 in the FOXP3 locus, thereby promoting FOXP3 transcription and TREG cell stability. This process, which is essential for TREG lineage stability and function, limits the severity of autoimmune neuroinflammation and infectious diseases, and favors tumor progression. These findings suggest that the regulation of intracellular iron by FTH is a stable property of TREG cells that supports immune homeostasis and limits the pathological outcomes of immune-mediated inflammation.
Keywords: FOXP3; Ferritin Heavy Chain; Iron Metabolism; Regulatory T Cells; Ten–eleven Translocation Enzymes.
© 2024. The Author(s).
Conflict of interest statement
MPS is a consultant to the New York Blood Center (NYBC), NYC, USA. The remaining authors declare no competing interests.
Figures











References
MeSH terms
Substances
Grants and funding
- RIGM 892773/EC | H2020 | PRIORITY 'Excellent science' | H2020 Marie Skłodowska-Curie Actions (MSCA)
- REGDAM 707998/EC | H2020 | PRIORITY 'Excellent science' | H2020 Marie Skłodowska-Curie Actions (MSCA)
- BILITOLERANCE 753236/EC | H2020 | PRIORITY 'Excellent science' | H2020 Marie Skłodowska-Curie Actions (MSCA)
- 20190090/International Postdoctoral Exchange Fellowship Program from the People's Republic of China
- 32171166/MOST | National Natural Science Foundation of China (NSFC)
- 82030003/MOST | National Natural Science Foundation of China (NSFC)
- SFRH/BPD/101608/2014/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- CEECIND/01589/2017/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- 2022.08590.PTDC/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- PTDC/MED-IMU/3649/2021/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- CEECIND/03106/2018/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- UID/Multi/04555/2013/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- SFRH/BPD/112135/2015/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- 283/BI/15/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- CONGENTO LISBOA-01-0145-FEDER-022170/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- 5723/2014/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- FEDER029411/MEC | Fundação para a Ciência e a Tecnologia (FCT)
- LT0043/2022-L/Human Frontier Science Program (HFSP)
- ALTF290-2017/European Molecular Biology Organization (EMBO)
- LCF/PR/HR22/52420023/'la Caixa' Foundation ('la Caixa')
- Excellence Cluster EXS 2051/Deutsche Forschungsgemeinschaft (DFG)
- SymbNET-Genomics and Metabolomics in a Host-Microbe Symbiosis Network Project -952537/EC | Horizon 2020 Framework Programme (H2020)
- BMBF 01EO1502/UKJ | Center for Sepsis Control and Care (CSCC)
- PID2019-105739GB-I00/MEC | Agencia Estatal de Investigación (AEI)
- LSBR 1818/Landsteiner Foundation for Blood Transfusion Research (LSBR)
- HR18-00502/la Caixa Foundation/FCT
- OPP1148170/Bill and Melinda Gates Foundation (GF)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

