SorLA restricts TNFα release from microglia to shape a glioma-supportive brain microenvironment
- PMID: 38499808
- PMCID: PMC11094098
- DOI: 10.1038/s44319-024-00117-6
SorLA restricts TNFα release from microglia to shape a glioma-supportive brain microenvironment
Abstract
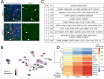
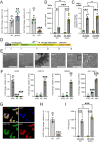
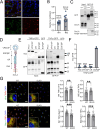
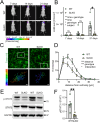
SorLA, encoded by the gene SORL1, is an intracellular sorting receptor of the VPS10P domain receptor gene family. Although SorLA is best recognized for its ability to shuttle target proteins between intracellular compartments in neurons, recent data suggest that also its microglial expression can be of high relevance for the pathogenesis of brain diseases, including glioblastoma (GBM). Here, we interrogated the impact of SorLA on the functional properties of glioma-associated microglia and macrophages (GAMs). In the GBM microenvironment, GAMs are re-programmed and lose the ability to elicit anti-tumor responses. Instead, they acquire a glioma-supporting phenotype, which is a key mechanism promoting glioma progression. Our re-analysis of published scRNA-seq data from GBM patients revealed that functional phenotypes of GAMs are linked to the level of SORL1 expression, which was further confirmed using in vitro models. Moreover, we demonstrate that SorLA restrains secretion of TNFα from microglia to restrict the inflammatory potential of these cells. Finally, we show that loss of SorLA exacerbates the pro-inflammatory response of microglia in the murine model of glioma and suppresses tumor growth.
Keywords: Brain Tumors; Glioblastoma; Intracellular Sorting; Phenotypic Polarization; VPS10P Domain Receptors.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Andersen OM, Reiche J, Schmidt V, Gotthardt M, Spoelgen R, Behlke J, von Arnim CAF, Breiderhoff T, Jansen P, Wu X, et al. Neuronal sorting protein-related receptor sorLA/LR11 regulates processing of the amyloid precursor protein. Proc Natl Acad Sci USA. 2005;102:13461–13466. doi: 10.1073/pnas.0503689102. - DOI - PMC - PubMed
-
- Caglayan S, Takagi-Niidome S, Liao F, Carlo A-S, Schmidt V, Burgert T, Kitago Y, Füchtbauer E-M, Füchtbauer A, Holtzman DM, et al. Lysosomal sorting of amyloid-β by the SORLA receptor is impaired by a familial Alzheimer’s disease mutation. Sci Transl Med. 2014;6:223ra20. doi: 10.1126/scitranslmed.3007747. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

