New plastids, old proteins: repeated endosymbiotic acquisitions in kareniacean dinoflagellates
- PMID: 38499810
- PMCID: PMC11014865
- DOI: 10.1038/s44319-024-00103-y
New plastids, old proteins: repeated endosymbiotic acquisitions in kareniacean dinoflagellates
Abstract
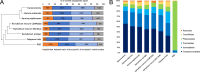
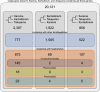
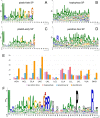
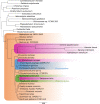
Dinoflagellates are a diverse group of ecologically significant micro-eukaryotes that can serve as a model system for plastid symbiogenesis due to their susceptibility to plastid loss and replacement via serial endosymbiosis. Kareniaceae harbor fucoxanthin-pigmented plastids instead of the ancestral peridinin-pigmented ones and support them with a diverse range of nucleus-encoded plastid-targeted proteins originating from the haptophyte endosymbiont, dinoflagellate host, and/or lateral gene transfers (LGT). Here, we present predicted plastid proteomes from seven distantly related kareniaceans in three genera (Karenia, Karlodinium, and Takayama) and analyze their evolutionary patterns using automated tree building and sorting. We project a relatively limited ( ~ 10%) haptophyte signal pointing towards a shared origin in the family Chrysochromulinaceae. Our data establish significant variations in the functional distributions of these signals, emphasizing the importance of micro-evolutionary processes in shaping the chimeric proteomes. Analysis of plastid genome sequences recontextualizes these results by a striking finding the extant kareniacean plastids are in fact not all of the same origin, as two of the studied species (Karlodinium armiger, Takayama helix) possess plastids from different haptophyte orders than the rest.
Keywords: Automated Tree Sorting; Myzozoa; Post-Endosymbiotic Organelle Evolution; Protists; Shopping Bag Model.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







Similar articles
-
Interpreting the complexities of the plastid genome in dinoflagellates: a mini-review of recent advances.Plant Mol Biol. 2024 Oct 21;114(6):114. doi: 10.1007/s11103-024-01511-3. Plant Mol Biol. 2024. PMID: 39432142 Review.
-
Phylogeny of nuclear-encoded plastid-targeted GAPDH gene supports separate origins for the peridinin- and the fucoxanthin derivative-containing plastids of dinoflagellates.Protist. 2004 Dec;155(4):447-58. doi: 10.1078/1434461042650325. Protist. 2004. PMID: 15648724
-
Patterns in evolutionary origins of heme, chlorophyll a and isopentenyl diphosphate biosynthetic pathways suggest non-photosynthetic periods prior to plastid replacements in dinoflagellates.PeerJ. 2018 Aug 3;6:e5345. doi: 10.7717/peerj.5345. eCollection 2018. PeerJ. 2018. PMID: 30083465 Free PMC article.
-
Fates of Evolutionarily Distinct, Plastid-type Glyceraldehyde 3-phosphate Dehydrogenase Genes in Kareniacean Dinoflagellates.J Eukaryot Microbiol. 2018 Jul;65(5):669-678. doi: 10.1111/jeu.12512. Epub 2018 Mar 15. J Eukaryot Microbiol. 2018. PMID: 29478272
-
Integration of plastids with their hosts: Lessons learned from dinoflagellates.Proc Natl Acad Sci U S A. 2015 Aug 18;112(33):10247-54. doi: 10.1073/pnas.1421380112. Epub 2015 May 20. Proc Natl Acad Sci U S A. 2015. PMID: 25995366 Free PMC article. Review.
Cited by
-
Interpreting the complexities of the plastid genome in dinoflagellates: a mini-review of recent advances.Plant Mol Biol. 2024 Oct 21;114(6):114. doi: 10.1007/s11103-024-01511-3. Plant Mol Biol. 2024. PMID: 39432142 Review.
-
Plastid translocon recycling in dinoflagellates demonstrates the portability of complex plastids between hosts.Curr Biol. 2024 Dec 2;34(23):5494-5506.e3. doi: 10.1016/j.cub.2024.10.034. Epub 2024 Nov 20. Curr Biol. 2024. PMID: 39571577 Free PMC article.
-
Prevalence and environmental abundance of the TSET complex in cosmopolitan algal groups.iScience. 2025 May 15;28(6):112679. doi: 10.1016/j.isci.2025.112679. eCollection 2025 Jun 20. iScience. 2025. PMID: 40524958 Free PMC article.
References
-
- Ardyna, M, D’Ovidio, F, Speich, S, Leconte, J, Chaffron, S, Audic, S, Garczarek, L, Pesant, S, Tara Oceans Consortium, C., & Tara Oceans Expedition, P. (2017). Environmental context of all samples from the Tara Oceans Expedition (2009-2013), about mesoscale features at the sampling location. In: Tara Oceans Consortium, Coordinators; Tara Oceans Expedition, Participants (2017): Registry of all samples from the Tara Oceans Expedition (2009-2013).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

