Whole genome identification, molecular docking and expression analysis of enzymes involved in the selenomethionine cycle in Cardamine hupingshanensis
- PMID: 38500044
- PMCID: PMC10949594
- DOI: 10.1186/s12870-024-04898-9
Whole genome identification, molecular docking and expression analysis of enzymes involved in the selenomethionine cycle in Cardamine hupingshanensis
Abstract
Background: The selenomethionine cycle (SeMTC) is a crucial pathway for the metabolism of selenium. The basic bioinformatics and functions of four enzymes involved in the cycle including S-adenosyl-methionine synthase (MAT), SAM-dependent methyltransferase (MTase), S-adenosyl-homocysteine hydrolase (SAHH) and methionine synthase (MTR), have been extensively reported in many eukaryotes. The identification and functional analyses of SeMTC genes/proteins in Cardamine hupingshanensis and their response to selenium stress have not yet been reported.
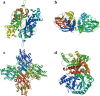
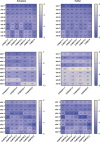
Results: In this study, 45 genes involved in SeMTC were identified in the C. hupingshanensis genome. Phylogenetic analysis showed that seven genes from ChMAT were clustered into four branches, twenty-seven genes from ChCOMT were clustered into two branches, four genes from ChSAHH were clustered into two branches, and seven genes from ChMTR were clustered into three branches. These genes were resided on 16 chromosomes. Gene structure and homologous protein modeling analysis illustrated that proteins in the same family are relatively conserved and have similar functions. Molecular docking showed that the affinity of SeMTC enzymes for selenium metabolites was higher than that for sulfur metabolites. The key active site residues identified for ChMAT were Ala269 and Lys273, while Leu221/231 and Gly207/249 were determined as the crucial residues for ChCOMT. For ChSAHH, the essential active site residues were found to be Asn87, Asp139 and Thr206/207/208/325. Ile204, Ser111/329/377, Asp70/206/254, and His329/332/380 were identified as the critical active site residues for ChMTR. In addition, the results of the expression levels of four enzymes under selenium stress revealed that ChMAT3-1 genes were upregulated approximately 18-fold, ChCOMT9-1 was upregulated approximately 38.7-fold, ChSAHH1-2 was upregulated approximately 11.6-fold, and ChMTR3-2 genes were upregulated approximately 28-fold. These verified that SeMTC enzymes were involved in response to selenium stress to varying degrees.
Conclusions: The results of this research are instrumental for further functional investigation of SeMTC in C. hupingshanensis. This also lays a solid foundation for deeper investigations into the physiological and biochemical mechanisms underlying selenium metabolism in plants.
Keywords: Cardamine Hupingshanensis; Gene expression; Molecular docking; Selenium stress response; Selenomethionine cycle.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
-
- O’Donohue B, Hiti-Bandaralage J, Gleeson M, O’Brien C, Harvey M-A, van der Ent A, Pinto Irish K, Mitter N, Hayward A. Tissue culture tools for selenium hyperaccumulator Neptunia Amplexicaulis for development in phytoextraction. Nat Prod Bioprospecting. 2022;12(1):28. doi: 10.1007/s13659-022-00351-2. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- B2020088/Scientific Research project of Hubei Provincial Education Department
- PT10202301/Open Foundation of Hubei Key Laboratory of Selenium Resource Research and Biological Application
- 32260070/National Natural Science Foundation of China
- T2020020/Excellent Young and Middle-aged Scientific and Technological Innovation Team Projects of Colleges and Universities in Hubei Province
LinkOut - more resources
Full Text Sources

