Development and validation a nomogram prediction model for early diagnosis of bloodstream infections in the intensive care unit
- PMID: 38500500
- PMCID: PMC10946253
- DOI: 10.3389/fcimb.2024.1348896
Development and validation a nomogram prediction model for early diagnosis of bloodstream infections in the intensive care unit
Abstract
Purpose: This study aims to develop and validate a nomogram for predicting the risk of bloodstream infections (BSI) in critically ill patients based on their admission status to the Intensive Care Unit (ICU).
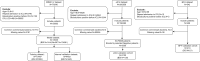
Patients and methods: Patients' data were extracted from the Medical Information Mart for Intensive Care-IV (MIMIC-IV) database (training set), the Beijing Friendship Hospital (BFH) database (validation set) and the eICU Collaborative Research Database (eICU-CRD) (validation set). Univariate logistic regression analyses were used to analyze the influencing factors, and lasso regression was used to select the predictive factors. Model performance was assessed using area under receiver operating characteristic curve (AUROC) and Presented as a Nomogram. Various aspects of the established predictive nomogram were evaluated, including discrimination, calibration, and clinical utility.
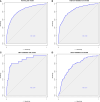
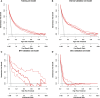
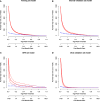
Results: The model dataset consisted of 14930 patients (1444 BSI patients) from the MIMIC-IV database, divided into the training and internal validation datasets in a 7:3 ratio. The eICU dataset included 2100 patients (100 with BSI) as the eICU validation dataset, and the BFH dataset included 419 patients (21 with BSI) as the BFH validation dataset. The nomogram was constructed based on Glasgow Coma Scale (GCS), sepsis related organ failure assessment (SOFA) score, temperature, heart rate, respiratory rate, white blood cell (WBC), red width of distribution (RDW), renal replacement therapy and presence of liver disease on their admission status to the ICU. The AUROCs were 0.83 (CI 95%:0.81-0.84) in the training dataset, 0.88 (CI 95%:0.88-0.96) in the BFH validation dataset, and 0.75 (95%CI 0.70-0.79) in the eICU validation dataset. The clinical effect curve and decision curve showed that most areas of the decision curve of this model were greater than 0, indicating that this model has a certain clinical effectiveness.
Conclusion: The nomogram developed in this study provides a valuable tool for clinicians and nurses to assess individual risk, enabling them to identify patients at a high risk of bloodstream infections in the ICU.
Keywords: bacteremia; bloodstream infections; critically ill; early diagnosis; intensive care unit; nomogram; prediction model.
Copyright © 2024 Qi, Dong, Lin and Duan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer ZZ declared a past co-authorship with the authors JL and MD to the handling editor.
Figures


Similar articles
-
Construction and evaluation of a mortality prediction model for patients with acute kidney injury undergoing continuous renal replacement therapy based on machine learning algorithms.Ann Med. 2024 Dec;56(1):2388709. doi: 10.1080/07853890.2024.2388709. Epub 2024 Aug 19. Ann Med. 2024. PMID: 39155811 Free PMC article.
-
[Development and validation of a prognostic model for patients with sepsis in intensive care unit].Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2023 Aug;35(8):800-806. doi: 10.3760/cma.j.cn121430-20230103-00003. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2023. PMID: 37593856 Chinese.
-
Nomogram predictive model for in-hospital mortality risk in elderly ICU patients with urosepsis.BMC Infect Dis. 2024 Apr 26;24(1):442. doi: 10.1186/s12879-024-09319-8. BMC Infect Dis. 2024. PMID: 38671376 Free PMC article.
-
The CMLA score: A novel tool for early prediction of renal replacement therapy in patients with cardiogenic shock.Curr Probl Cardiol. 2024 Dec;49(12):102870. doi: 10.1016/j.cpcardiol.2024.102870. Epub 2024 Sep 27. Curr Probl Cardiol. 2024. PMID: 39343053 Review.
-
Contribution of Open Access Databases to Intensive Care Medicine Research: Scoping Review.J Med Internet Res. 2025 Jan 9;27:e57263. doi: 10.2196/57263. J Med Internet Res. 2025. PMID: 39787600 Free PMC article.
Cited by
-
Development and application of an early prediction model for risk of bloodstream infection based on real-world study.BMC Med Inform Decis Mak. 2025 May 14;25(1):186. doi: 10.1186/s12911-025-03020-9. BMC Med Inform Decis Mak. 2025. PMID: 40369550 Free PMC article.
-
An externally validated prognostic model for critically ill patients with traumatic brain injury.Ann Clin Transl Neurol. 2024 Sep;11(9):2350-2359. doi: 10.1002/acn3.52148. Epub 2024 Jul 7. Ann Clin Transl Neurol. 2024. PMID: 38973122 Free PMC article.
-
Clinical features and risk factors of older adults with bloodstream infection.BMC Geriatr. 2025 May 31;25(1):397. doi: 10.1186/s12877-025-05934-5. BMC Geriatr. 2025. PMID: 40450236 Free PMC article.
References
-
- Esbenshade A. J., Zhao Z., Baird A., Holmes E. A., Dulek D. E., Banerjee R., et al. . (2020). Prospective implementation of a risk prediction model for bloodstream infection safely reduces antibiotic usage in febrile pediatric cancer patients without severe neutropenia. J. Clin. Oncol. 38, 3150–3160. doi: 10.1200/JCO.20.00591 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

