Design of AsLOV2 domain as a carrier of light-induced dissociable FMN photosensitizer
- PMID: 38501448
- PMCID: PMC10949324
- DOI: 10.1002/pro.4921
Design of AsLOV2 domain as a carrier of light-induced dissociable FMN photosensitizer
Abstract
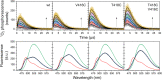
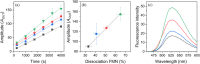
Flavin mononucleotide (FMN) is a highly efficient photosensitizer (PS) yielding singlet oxygen (1 O2 ). However, its 1 O2 production efficiency significantly decreases upon isoalloxazine ring encapsulation into the protein matrix in genetically encoded photosensitizers (GEPS). Reducing isoalloxazine ring interactions with surrounding amino acids by protein engineering may increase 1 O2 production efficiency GEPS, but at the same time weakened native FMN-protein interactions may cause undesirable FMN dissociation. Here, in contrast, we intentionally induce the FMN release by light-triggered sulfur oxidation of strategically placed cysteines (oxidation-prone amino acids) in the isoalloxazine-binding site due to significantly increased volume of the cysteinyl side residue(s). As a proof of concept, in three variants of the LOV2 domain of Avena sativa (AsLOV2), namely V416C, T418C, and V416C/T418C, the effective 1 O2 production strongly correlated with the efficiency of irradiation-induced FMN dissociation (wild type (WT) < V416C < T418C < V416C/T418C). This alternative approach enables us: (i) to overcome the low 1 O2 production efficiency of flavin-based GEPSs without affecting native isoalloxazine ring-protein interactions and (ii) to utilize AsLOV2, due to its inherent binding propensity to FMN, as a PS vehicle, which is released at a target by light irradiation.
Keywords: LOV2 domain; flavin cofactor; genetically encoded photosensitizers; miniSOG; singlet oxygen.
© 2024 The Protein Society.
Figures



References
-
- Andersen HC. RATTLE – a velocity version of the SHAKE algorithms for molecular‐dynamics calculations. J Comput Phys. 1983;52(1):24–34. 10.1016/0021-9991(83)90014-1 - DOI
-
- Darden T, York D, Pedersen L. Particle mesh Ewald – an N.Log(N) method for Ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. 10.1063/1.464397 - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

