A cellulosomal double-dockerin module from Clostridium thermocellum shows distinct structural and cohesin-binding features
- PMID: 38501488
- PMCID: PMC10949318
- DOI: 10.1002/pro.4937
A cellulosomal double-dockerin module from Clostridium thermocellum shows distinct structural and cohesin-binding features
Abstract
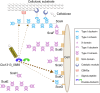
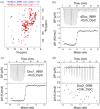
Cellulosomes are intricate cellulose-degrading multi-enzymatic complexes produced by anaerobic bacteria, which are valuable for bioenergy development and biotechnology. Cellulosome assembly relies on the selective interaction between cohesin modules in structural scaffolding proteins (scaffoldins) and dockerin modules in enzymes. Although the number of tandem cohesins in the scaffoldins is believed to determine the complexity of the cellulosomes, tandem dockerins also exist, albeit very rare, in some cellulosomal components whose assembly and functional roles are currently unclear. In this study, we characterized the structure and mode of assembly of a tandem bimodular double-dockerin, which is connected to a putative S8 protease in the cellulosome-producing bacterium, Clostridium thermocellum. Crystal and NMR structures of the double-dockerin revealed two typical type I dockerin folds with significant interactions between them. Interaction analysis by isothermal titration calorimetry and NMR titration experiments revealed that the double-dockerin displays a preference for binding to the cell-wall anchoring scaffoldin ScaD through the first dockerin with a canonical dual-binding mode, while the second dockerin module was unable to bind to any of the tested cohesins. Surprisingly, the double-dockerin showed a much higher affinity to a cohesin from the CipC scaffoldin of Clostridium cellulolyticum than to the resident cohesins from C. thermocellum. These results contribute valuable insights into the structure and assembly of the double-dockerin module, and provide the basis for further functional studies on multiple-dockerin modules and cellulosomal proteases, thus highlighting the complexity and diversity of cellulosomal components.
Keywords: NMR; X-ray crystallography; binding affinity; protein complex; protein-protein interaction; scaffolding protein; tandem modules.
© 2024 The Protein Society.
Conflict of interest statement
There are no conflicts of interest to declare.
Figures




References
MeSH terms
Substances
Grants and funding
- 2023YFC3402300/National Key Research and Development Program of China
- 32070125/National Natural Science Foundation of China
- 32200030/National Natural Science Foundation of China
- 32170051/National Natural Science Foundation of China
- 32171203/National Natural Science Foundation of China
- 32070028/National Natural Science Foundation of China
- ICP202304/QIBEBT International Cooperation Project
- M2022-01/State Key Laboratory of Microbial Technology Open Projects Fund
- ZR2016CB09/Shandong Provincial Natural Science Foundation
- FRF-DF-20-09/Fundamental Research Funds for the Central Universities
- 2302020JXGGRC-005/Training Program for Young Teaching Backbone Talents
- JG2021ZD01/Major Education and Teaching Reform Research Project
LinkOut - more resources
Full Text Sources

