Functional dissection of the spike glycoprotein S1 subunit and identification of cellular cofactors for regulation of swine acute diarrhea syndrome coronavirus entry
- PMID: 38501663
- PMCID: PMC11019839
- DOI: 10.1128/jvi.00139-24
Functional dissection of the spike glycoprotein S1 subunit and identification of cellular cofactors for regulation of swine acute diarrhea syndrome coronavirus entry
Abstract
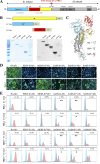
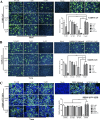
Swine acute diarrhea syndrome coronavirus (SADS-CoV) is a novel porcine enteric coronavirus, and the broad interspecies infection of SADS-CoV poses a potential threat to human health. This study provides experimental evidence to dissect the roles of distinct domains within the SADS-CoV spike S1 subunit in cellular entry. Specifically, we expressed the S1 and its subdomains, S1A and S1B. Cell binding and invasion inhibition assays revealed a preference for the S1B subdomain in binding to the receptors on the cell surface, and this unknown receptor is not utilized by the porcine epidemic diarrhea virus. Nanoparticle display demonstrated hemagglutination of erythrocytes from pigs, humans, and mice, linking the S1A subdomain to the binding of sialic acid (Sia) involved in virus attachment. We successfully rescued GFP-labeled SADS-CoV (rSADS-GFP) from a recombinant cDNA clone to track viral infection. Antisera raised against S1, S1A, or S1B contained highly potent neutralizing antibodies, with anti-S1B showing better efficiency in neutralizing rSADS-GFP infection compared to anti-S1A. Furthermore, depletion of heparan sulfate (HS) by heparinase treatment or pre-incubation of rSADS-GFP with HS or constituent monosaccharides could inhibit SADS-CoV entry. Finally, we demonstrated that active furin cleavage of S glycoprotein and the presence of type II transmembrane serine protease (TMPRSS2) are essential for SADS-CoV infection. These combined observations suggest that the wide cell tropism of SADS-CoV may be related to the distribution of Sia or HS on the cell surface, whereas the S1B contains the main protein receptor binding site. Specific host proteases also play important roles in facilitating SADS-CoV entry.IMPORTANCESwine acute diarrhea syndrome coronavirus (SADS-CoV) is a novel pathogen infecting piglet, and its unique genetic evolution characteristics and broad species tropism suggest the potential for cross-species transmission. The virus enters cells through its spike (S) glycoprotein. In this study, we identify the receptor binding domain on the C-terminal part of the S1 subunit (S1B) of SADS-CoV, whereas the sugar-binding domain located at the S1 N-terminal part of S1 (S1A). Sialic acid, heparan sulfate, and specific host proteases play essential roles in viral attachment and entry. The dissection of SADS-CoV S1 subunit's functional domains and identification of cellular entry cofactors will help to explore the receptors used by SADS-CoV, which may contribute to exploring the mechanisms behind cross-species transmission and host tropism.
Keywords: TMPRSS2; heparan sulfate; receptor-binding domain (RBD); sialic acid; spike; swine acute diarrhea syndrome coronavirus (SADS-CoV).
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Cryo-electron Microscopy Structure of the Swine Acute Diarrhea Syndrome Coronavirus Spike Glycoprotein Provides Insights into Evolution of Unique Coronavirus Spike Proteins.J Virol. 2020 Oct 27;94(22):e01301-20. doi: 10.1128/JVI.01301-20. Print 2020 Oct 27. J Virol. 2020. PMID: 32817223 Free PMC article.
-
Broad Cross-Species Infection of Cultured Cells by Bat HKU2-Related Swine Acute Diarrhea Syndrome Coronavirus and Identification of Its Replication in Murine Dendritic Cells In Vivo Highlight Its Potential for Diverse Interspecies Transmission.J Virol. 2019 Nov 26;93(24):e01448-19. doi: 10.1128/JVI.01448-19. Print 2019 Dec 15. J Virol. 2019. PMID: 31554686 Free PMC article.
-
Establishment of replication-competent vesicular stomatitis virus recapitulating SADS-CoV entry.J Virol. 2024 May 14;98(5):e0195723. doi: 10.1128/jvi.01957-23. Epub 2024 Apr 1. J Virol. 2024. PMID: 38557247 Free PMC article.
-
Three Main Inducers of Alphacoronavirus Infection of Enterocytes: Sialic Acid, Proteases, and Low pH.Intervirology. 2018;61(2):53-63. doi: 10.1159/000492424. Epub 2018 Sep 3. Intervirology. 2018. PMID: 30176660 Free PMC article. Review.
-
Cellular entry of the porcine epidemic diarrhea virus.Virus Res. 2016 Dec 2;226:117-127. doi: 10.1016/j.virusres.2016.05.031. Epub 2016 Jun 15. Virus Res. 2016. PMID: 27317167 Free PMC article. Review.
Cited by
-
Interplay of swine acute diarrhoea syndrome coronavirus and the host intrinsic and innate immunity.Vet Res. 2025 Jan 9;56(1):5. doi: 10.1186/s13567-024-01436-1. Vet Res. 2025. PMID: 39789633 Free PMC article. Review.
-
Swine Acute Diarrhea Syndrome Coronavirus: An Overview of Virus Structure and Virus-Host Interactions.Animals (Basel). 2025 Jan 9;15(2):149. doi: 10.3390/ani15020149. Animals (Basel). 2025. PMID: 39858149 Free PMC article. Review.
References
-
- Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, Hiller M, Koepfli KP, Pfenning AR, Zhao H, Genereux DP, Swofford R, Pollard KS, Ryder OA, Nweeia MT, Lindblad-Toh K, Teeling EC, Karlsson EK, Lewin HA. 2020. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci U S A 117:22311–22322. doi:10.1073/pnas.2010146117 - DOI - PMC - PubMed
-
- Starr TN, Greaney AJ, Hilton SK, Ellis D, Crawford KHD, Dingens AS, Navarro MJ, Bowen JE, Tortorici MA, Walls AC, King NP, Veesler D, Bloom JD. 2020. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 182:1295–1310. doi:10.1016/j.cell.2020.08.012 - DOI - PMC - PubMed
-
- Hodcroft EB, Zuber M, Nadeau S, Vaughan TG, Crawford KHD, Althaus CL, Reichmuth ML, Bowen JE, Walls AC, Corti D, Bloom JD, Veesler D, Mateo D, Hernando A, Comas I, González-Candelas F, Stadler T, Neher RA, SeqCOVID-SPAIN consortium . 2021. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature 595:707–712. doi:10.1038/s41586-021-03677-y - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

