OsbZIP18 Is a Positive Regulator of Phenylpropanoid and Flavonoid Biosynthesis under UV-B Radiation in Rice
- PMID: 38502046
- PMCID: PMC10893026
- DOI: 10.3390/plants13040498
OsbZIP18 Is a Positive Regulator of Phenylpropanoid and Flavonoid Biosynthesis under UV-B Radiation in Rice
Abstract
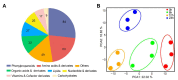
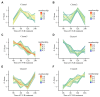
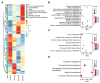
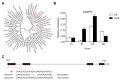
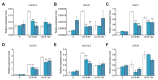
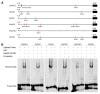
In plants exposed to ultraviolet B radiation (UV-B; 280-315 nm), metabolic responses are activated, which reduce the damage caused by UV-B. Although several metabolites responding to UV-B stress have been identified in plants, the accumulation of these metabolites at different time points under UV-B stress remains largely unclear, and the transcription factors regulating these metabolites have not been well characterized. Here, we explored the changes in metabolites in rice after UV-B treatment for 0 h, 6 h, 12 h, and 24 h and identified six patterns of metabolic change. We show that the rice transcription factor OsbZIP18 plays an important role in regulating phenylpropanoid and flavonoid biosynthesis under UV-B stress in rice. Metabolic profiling revealed that the contents of phenylpropanoid and flavonoid were significantly reduced in osbzip18 mutants compared with the wild-type plants (WT) under UV-B stress. Further analysis showed that the expression of many genes involved in the phenylpropanoid and flavonoid biosynthesis pathways was lower in osbzip18 mutants than in WT plants, including OsPAL5, OsC4H, Os4CL, OsCHS, OsCHIL2, and OsF3H. Electrophoretic mobility shift assays (EMSA) revealed that OsbZIP18 bind to the promoters of these genes, suggesting that OsbZIP18 function is an important positive regulator of phenylpropanoid and flavonoid biosynthesis under UV-B stress. In conclusion, our findings revealed that OsbZIP18 is an essential regulator for phenylpropanoid and flavonoid biosynthesis and plays a crucial role in regulating UV-B stress responses in rice.
Keywords: UV-B radiation; metabolites; phenylpropanoid and flavonoids biosynthesis; regulator; rice.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
OsbZIP18, a Positive Regulator of Serotonin Biosynthesis, Negatively Controls the UV-B Tolerance in Rice.Int J Mol Sci. 2022 Mar 16;23(6):3215. doi: 10.3390/ijms23063215. Int J Mol Sci. 2022. PMID: 35328636 Free PMC article.
-
UV-B induces the expression of flavonoid biosynthetic pathways in blueberry (Vaccinium corymbosum) calli.Front Plant Sci. 2022 Nov 22;13:1079087. doi: 10.3389/fpls.2022.1079087. eCollection 2022. Front Plant Sci. 2022. PMID: 36483950 Free PMC article.
-
Natural variation in the OsbZIP18 promoter contributes to branched-chain amino acid levels in rice.New Phytol. 2020 Dec;228(5):1548-1558. doi: 10.1111/nph.16800. Epub 2020 Aug 6. New Phytol. 2020. PMID: 32654152
-
Glycosylation Is a Major Regulator of Phenylpropanoid Availability and Biological Activity in Plants.Front Plant Sci. 2016 May 26;7:735. doi: 10.3389/fpls.2016.00735. eCollection 2016. Front Plant Sci. 2016. PMID: 27303427 Free PMC article. Review.
-
MYB transcription factors-master regulators of phenylpropanoid biosynthesis and diverse developmental and stress responses.Plant Cell Rep. 2022 Dec;41(12):2245-2260. doi: 10.1007/s00299-022-02927-1. Epub 2022 Sep 29. Plant Cell Rep. 2022. PMID: 36171500 Review.
Cited by
-
Comprehensive Modulation of Secondary Metabolites in Terpenoid-Accumulating Mentha spicata L. via UV Radiation.Plants (Basel). 2024 Jun 24;13(13):1746. doi: 10.3390/plants13131746. Plants (Basel). 2024. PMID: 38999586 Free PMC article.
-
Phenylpropanoids metabolism: recent insight into stress tolerance and plant development cues.Front Plant Sci. 2025 Jun 26;16:1571825. doi: 10.3389/fpls.2025.1571825. eCollection 2025. Front Plant Sci. 2025. PMID: 40641862 Free PMC article. Review.
-
Metabolomic Analysis of Elymus sibiricus Exposed to UV-B Radiation Stress.Molecules. 2024 Oct 30;29(21):5133. doi: 10.3390/molecules29215133. Molecules. 2024. PMID: 39519780 Free PMC article.
-
Enhanced Synthesis of Volatile Compounds by UV-B Irradiation in Artemisia argyi Leaves.Metabolites. 2024 Dec 11;14(12):700. doi: 10.3390/metabo14120700. Metabolites. 2024. PMID: 39728481 Free PMC article.
-
Genetic Diversity and Metabolic Profile of Tibetan Medicinal Plant Saussurea obvallata.Genes (Basel). 2025 May 17;16(5):593. doi: 10.3390/genes16050593. Genes (Basel). 2025. PMID: 40428415 Free PMC article.
References
-
- Mohammed A.R., Rounds E.W., Tarpley L. Response of rice (Oryza sativa L.) tillering to sub-ambient levels of Ultraviolet-B radiation. J. Agron. Crop Sci. 2010;193:324–335. doi: 10.1111/j.1439-037X.2007.00268.x. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

