SMARCAL1 ubiquitylation controls its association with RPA-coated ssDNA and promotes replication fork stability
- PMID: 38502677
- PMCID: PMC10950228
- DOI: 10.1371/journal.pbio.3002552
SMARCAL1 ubiquitylation controls its association with RPA-coated ssDNA and promotes replication fork stability
Abstract
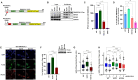
Impediments in replication fork progression cause genomic instability, mutagenesis, and severe pathologies. At stalled forks, RPA-coated single-stranded DNA (ssDNA) activates the ATR kinase and directs fork remodeling, 2 key early events of the replication stress response. RFWD3, a recently described Fanconi anemia (FA) ubiquitin ligase, associates with RPA and promotes its ubiquitylation, facilitating late steps of homologous recombination (HR). Intriguingly, RFWD3 also regulates fork progression, restart and stability via poorly understood mechanisms. Here, we used proteomics to identify putative RFWD3 substrates during replication stress in human cells. We show that RFWD3 interacts with and ubiquitylates the SMARCAL1 DNA translocase directly in vitro and following DNA damage in vivo. SMARCAL1 ubiquitylation does not trigger its subsequent proteasomal degradation but instead disengages it from RPA thereby regulating its function at replication forks. Proper regulation of SMARCAL1 by RFWD3 at stalled forks protects them from excessive MUS81-mediated cleavage in response to UV irradiation, thereby limiting DNA replication stress. Collectively, our results identify RFWD3-mediated SMARCAL1 ubiquitylation as a novel mechanism that modulates fork remodeling to avoid genome instability triggered by aberrant fork processing.
Copyright: © 2024 Yates et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

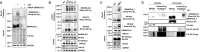




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

