Vulnerability to APOBEC3G linked to the pathogenicity of deltaretroviruses
- PMID: 38502701
- PMCID: PMC10990082
- DOI: 10.1073/pnas.2309925121
Vulnerability to APOBEC3G linked to the pathogenicity of deltaretroviruses
Abstract
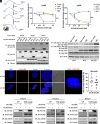
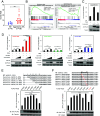
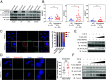
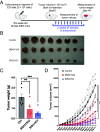
Human retroviruses are derived from simian ones through cross-species transmission. These retroviruses are associated with little pathogenicity in their natural hosts, but in humans, HIV causes AIDS, and human T-cell leukemia virus type 1 (HTLV-1) induces adult T-cell leukemia-lymphoma (ATL). We analyzed the proviral sequences of HTLV-1, HTLV-2, and simian T-cell leukemia virus type 1 (STLV-1) from Japanese macaques (Macaca fuscata) and found that APOBEC3G (A3G) frequently generates G-to-A mutations in the HTLV-1 provirus, whereas such mutations are rare in the HTLV-2 and STLV-1 proviruses. Therefore, we investigated the mechanism of how HTLV-2 is resistant to human A3G (hA3G). HTLV-1, HTLV-2, and STLV-1 encode the so-called antisense proteins, HTLV-1 bZIP factor (HBZ), Antisense protein of HTLV-2 (APH-2), and STLV-1 bZIP factor (SBZ), respectively. APH-2 efficiently inhibits the deaminase activity of both hA3G and simian A3G (sA3G). HBZ and SBZ strongly suppress sA3G activity but only weakly inhibit hA3G, suggesting that HTLV-1 is incompletely adapted to humans. Unexpectedly, hA3G augments the activation of the transforming growth factor (TGF)-β/Smad pathway by HBZ, and this activation is associated with ATL cell proliferation by up-regulating BATF3/IRF4 and MYC. In contrast, the combination of APH-2 and hA3G, or the combination of SBZ and sA3G, does not enhance the TGF-β/Smad pathway. Thus, HTLV-1 is vulnerable to hA3G but utilizes it to promote the proliferation of infected cells via the activation of the TGF-β/Smad pathway. Antisense factors in each virus, differently adapted to control host cellular functions through A3G, seem to dictate the pathogenesis.
Keywords: APOBEC3G; HBZ; HTLV-1; TGF-β; deltaretrovirus.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures


References
-
- Kalyanaraman V. S., et al. , A new subtype of human T-cell leukemia virus (HTLV-II) associated with a T-cell variant of hairy cell leukemia. Science 218, 571–573 (1982). - PubMed
-
- Barre-Sinoussi F., et al. , Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220, 868–871 (1983). - PubMed
-
- Barin F., et al. , Serological evidence for virus related to simian T-lymphotropic retrovirus III in residents of west Africa. Lancet 2, 1387–1389 (1985). - PubMed
-
- Kanki P. J., et al. , New human T-lymphotropic retrovirus related to simian T-lymphotropic virus type III (STLV-IIIAGM). Science 232, 238–243 (1986). - PubMed

