Morc1 reestablishes H3K9me3 heterochromatin on piRNA-targeted transposons in gonocytes
- PMID: 38502704
- PMCID: PMC10990106
- DOI: 10.1073/pnas.2317095121
Morc1 reestablishes H3K9me3 heterochromatin on piRNA-targeted transposons in gonocytes
Abstract
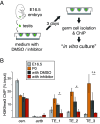
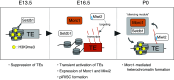
To maintain fertility, male mice re-repress transposable elements (TEs) that were de-silenced in the early gonocytes before their differentiation into spermatogonia. However, the mechanism of TE silencing re-establishment remains unknown. Here, we found that the DNA-binding protein Morc1, in cooperation with the methyltransferase SetDB1, deposits the repressive histone mark H3K9me3 on a large fraction of activated TEs, leading to heterochromatin. Morc1 also triggers DNA methylation, but TEs targeted by Morc1-driven DNA methylation only slightly overlapped with those repressed by Morc1/SetDB1-dependent heterochromatin formation, suggesting that Morc1 silences TEs in two different manners. In contrast, TEs regulated by Morc1 and Miwi2, the nuclear PIWI-family protein, almost overlapped. Miwi2 binds to PIWI-interacting RNAs (piRNAs) that base-pair with TE mRNAs via sequence complementarity, while Morc1 DNA binding is not sequence specific, suggesting that Miwi2 selects its targets, and then, Morc1 acts to repress them with cofactors. A high-ordered mechanism of TE repression in gonocytes has been identified.
Keywords: Morc1; gonocyte; heterochromatin; piRNA.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures





References
-
- Fedoroff N. V., Transposable elements, epigenetics, and genome evolution. Science 338, 758–767 (2012). - PubMed
-
- Bourc'his D., Xu G. L., Lin C. S., Bollman B., Bestor T. H., Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001). - PubMed
-
- Bourc’his D., Bestor T. H., Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99 (2004). - PubMed
-
- Sasaki H., Matsui Y., Epigenetic events in mammalian germ-cell development: Reprogramming and beyond. Nat. Rev. Genet. 9, 129–140 (2008). - PubMed
-
- Seki Y., et al. , Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev. Biol. 278, 440–458 (2005). - PubMed
MeSH terms
Substances
Grants and funding
- 21bm0704041h0003/Japan Agency for Medical Research and Development (AMED)
- JP19H05466/Ministry of Education, Culture, Sports, Science and Technology (MEXT)
- 22K06338/MEXT | Japan Society for the Promotion of Science (JSPS)
- 19K06616/MEXT | Japan Society for the Promotion of Science (JSPS)
- 22jm0210084h0003/Japan Agency for Medical Research and Development (AMED)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

