Cigarette Smoking and E-cigarette Use Induce Shared DNA Methylation Changes Linked to Carcinogenesis
- PMID: 38503267
- PMCID: PMC11148547
- DOI: 10.1158/0008-5472.CAN-23-2957
Cigarette Smoking and E-cigarette Use Induce Shared DNA Methylation Changes Linked to Carcinogenesis
Abstract
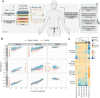
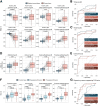
Tobacco use is a major modifiable risk factor for adverse health outcomes, including cancer, and elicits profound epigenetic changes thought to be associated with long-term cancer risk. While electronic cigarettes (e-cigarettes) have been advocated as harm reduction alternatives to tobacco products, recent studies have revealed potential detrimental effects, highlighting the urgent need for further research into the molecular and health impacts of e-cigarettes. Here, we applied computational deconvolution methods to dissect the cell- and tissue-specific epigenetic effects of tobacco or e-cigarette use on DNA methylation (DNAme) in over 3,500 buccal/saliva, cervical, or blood samples, spanning epithelial and immune cells at directly and indirectly exposed sites. The 535 identified smoking-related DNAme loci [cytosine-phosphate-guanine sites (CpG)] clustered into four functional groups, including detoxification or growth signaling, based on cell type and anatomic site. Loci hypermethylated in buccal epithelial cells of smokers associated with NOTCH1/RUNX3/growth factor receptor signaling also exhibited elevated methylation in cancer tissue and progressing lung carcinoma in situ lesions, and hypermethylation of these sites predicted lung cancer development in buccal samples collected from smokers up to 22 years prior to diagnosis, suggesting a potential role in driving carcinogenesis. Alarmingly, these CpGs were also hypermethylated in e-cigarette users with a limited smoking history. This study sheds light on the cell type-specific changes to the epigenetic landscape induced by smoking-related products.
Significance: The use of both cigarettes and e-cigarettes elicits cell- and exposure-specific epigenetic effects that are predictive of carcinogenesis, suggesting caution when broadly recommending e-cigarettes as aids for smoking cessation.
©2024 The Authors; Published by the American Association for Cancer Research.
Figures





Comment in
-
Asia's Teen Vaping Surge: Unmasking Risks and Mobilizing Solutions.Asia Pac J Public Health. 2024 Sep;36(6-7):646-647. doi: 10.1177/10105395241273180. Epub 2024 Aug 19. Asia Pac J Public Health. 2024. PMID: 39158033 No abstract available.
References
-
- United States Public Health Service Office of the Surgeon General, Centers for Disease Control and Prevention (U.S.), National Center for Chronic Disease Prevention and Health Promotion (U.S.) Office on Smoking and Health. How tobacco smoke causes disease: the biology and behavioral basis for smoking-attributable disease : a report of the surgeon general. Atlanta (GA): Centers for Disease Control and Prevention; 2010. - PubMed
-
- Lushniak BD, Samet JM, Pechacek TF, Norman LA, Taylor PA. The health consequences of smoking—50 years of progress : a report of the surgeon general. Atlanta (GA): Centers for Disease Control and Prevention; 2014.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

