Seedling microbiota engineering using bacterial synthetic community inoculation on seeds
- PMID: 38503562
- PMCID: PMC10977042
- DOI: 10.1093/femsec/fiae027
Seedling microbiota engineering using bacterial synthetic community inoculation on seeds
Erratum in
-
Correction to: Seedling microbiota engineering using bacterial synthetic community inoculation on seeds.FEMS Microbiol Ecol. 2024 May 14;100(6):fiae065. doi: 10.1093/femsec/fiae065. FEMS Microbiol Ecol. 2024. PMID: 38748655 Free PMC article. No abstract available.
Abstract
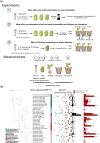
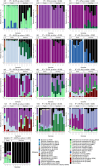
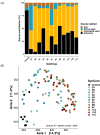
Synthetic Communities (SynComs) are being developed and tested to manipulate plant microbiota and improve plant health. To date, only few studies proposed the use of SynCom on seed despite its potential for plant microbiota engineering. We developed and presented a simple and effective seedling microbiota engineering method using SynCom inoculation on seeds. The method was successful using a wide diversity of SynCom compositions and bacterial strains that are representative of the common bean seed microbiota. First, this method enables the modulation of seed microbiota composition and community size. Then, SynComs strongly outcompeted native seed and potting soil microbiota and contributed on average to 80% of the seedling microbiota. We showed that strain abundance on seed was a main driver of an effective seedling microbiota colonization. Also, selection was partly involved in seed and seedling colonization capacities since strains affiliated to Enterobacteriaceae and Erwiniaceae were good colonizers while Bacillaceae and Microbacteriaceae were poor colonizers. Additionally, the engineered seed microbiota modified the recruitment and assembly of seedling and rhizosphere microbiota through priority effects. This study shows that SynCom inoculation on seeds represents a promising approach to study plant microbiota assembly and its consequence on plant fitness.
Keywords: Synthetic Community; microbiota engineering; seed microbiota; seedling microbiota; transmission.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures