ESI-MS analysis of Cu(I) binding to apo and Zn7 human metallothionein 1A, 2, and 3 identifies the formation of a similar series of metallated species with no individual isoform optimization for Cu(I)
- PMID: 38503570
- PMCID: PMC11004924
- DOI: 10.1093/mtomcs/mfae015
ESI-MS analysis of Cu(I) binding to apo and Zn7 human metallothionein 1A, 2, and 3 identifies the formation of a similar series of metallated species with no individual isoform optimization for Cu(I)
Abstract
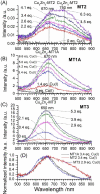
Metallothioneins (MTs) are cysteine-rich proteins involved in metal homeostasis, heavy metal detoxification, and protection against oxidative stress. Whether the four mammalian MT isoforms exhibit different metal binding properties is not clear. In this paper, the Cu(I) binding properties of the apo MT1A, apo MT2, and apo MT3 are compared and the relative Cu(I) binding affinities are reported. In all three isoforms, Cu4, Cu6, and Cu10 species form cooperatively, and MT1A and MT2 also form a Cu13 species. The Cu(I) binding properties of Zn7-MT1A, Zn7-MT2, and Zn7-MT3 are compared systematically using isotopically pure 63Cu(I) and 68Zn(II). The species formed in each MT isoform were detected through electrospray ionization-mass spectrometry and further characterized using room temperature phosphorescence spectroscopy. The mixed metal Cu, Zn species forming in MT1A, MT2, and MT3 have similar stoichiometries and their emission spectral properties indicate that analogous clusters form in the three isoforms. Three parallel metallation pathways have been proposed through analysis of the detailed Cu, Zn speciation in MT1A, MT2, and MT3. Pathway ① results in Cu5Zn5-MT and Cu9Zn3-MT. Pathway ② involves Cu6Zn4-MT and Cu10Zn2-MT. Pathway ③ includes Cu8Zn4-MT. Speciation analysis indicates that Pathway ② is the preferred pathway for MT2. This is also evident in the phosphorescence spectra with the 750 nm emission from Cu6Zn4-MT being most prominent in MT2. We see no evidence for different MT isoforms being optimized or exhibiting preferences for certain metals. We discuss the probable stoichiometry for MTs in vivo based on the in vitro determined binding constants.
Keywords: Cu homeostasis; Cu-thiolate clusters; ESI–MS; Zn homeostasis; metallothionein; phosphorescence.
© The Author(s) 2024. Published by Oxford University Press.
Conflict of interest statement
There are no conflicts of interest.
Figures














References
-
- Robbins A., McRee D., Williamson M., Collett S., Xuong N., Furey W., Wang B., Stout C., Refined crystal structure of Cd, Zn metallothionein at 2.0 Åresolution, J. Mol. Biol., 1991, 221(4), 1269–1293. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

