VCF1 is a p97/VCP cofactor promoting recognition of ubiquitylated p97-UFD1-NPL4 substrates
- PMID: 38503733
- PMCID: PMC10950897
- DOI: 10.1038/s41467-024-46760-4
VCF1 is a p97/VCP cofactor promoting recognition of ubiquitylated p97-UFD1-NPL4 substrates
Abstract
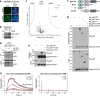
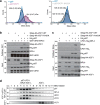
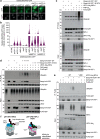
The hexameric AAA+ ATPase p97/VCP functions as an essential mediator of ubiquitin-dependent cellular processes, extracting ubiquitylated proteins from macromolecular complexes or membranes by catalyzing their unfolding. p97 is directed to ubiquitylated client proteins via multiple cofactors, most of which interact with the p97 N-domain. Here, we discover that FAM104A, a protein of unknown function also named VCF1 (VCP/p97 nuclear Cofactor Family member 1), acts as a p97 cofactor in human cells. Detailed structure-function studies reveal that VCF1 directly binds p97 via a conserved α-helical motif that recognizes the p97 N-domain with unusually high affinity, exceeding that of other cofactors. We show that VCF1 engages in joint p97 complex formation with the heterodimeric primary p97 cofactor UFD1-NPL4 and promotes p97-UFD1-NPL4-dependent proteasomal degradation of ubiquitylated substrates in cells. Mechanistically, VCF1 indirectly stimulates UFD1-NPL4 interactions with ubiquitin conjugates via its binding to p97 but has no intrinsic affinity for ubiquitin. Collectively, our findings establish VCF1 as an unconventional p97 cofactor that promotes p97-dependent protein turnover by facilitating p97-UFD1-NPL4 recruitment to ubiquitylated targets.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
- NNF14CC0001/Novo Nordisk Fonden (Novo Nordisk Foundation)
- NNF18OC0030752/Novo Nordisk Fonden (Novo Nordisk Foundation)
- R223-2016-281/Lundbeckfonden (Lundbeck Foundation)
- 0134-00048B/Det Frie Forskningsråd (Danish Council for Independent Research)
- DNRF-115/Danmarks Grundforskningsfond (Danish National Research Foundation)
- 812829 (aDDRess)/EC | Horizon 2020 Framework Programme (EU Framework Programme for Research and Innovation H2020)
- ALTF 1149-2020/European Molecular Biology Organization (EMBO)
- 424228829/Deutsche Forschungsgemeinschaft (German Research Foundation)
- 715975/EC | EU Framework Programme for Research and Innovation H2020 | H2020 Priority Excellent Science | H2020 European Research Council (H2020 Excellent Science - European Research Council)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

